
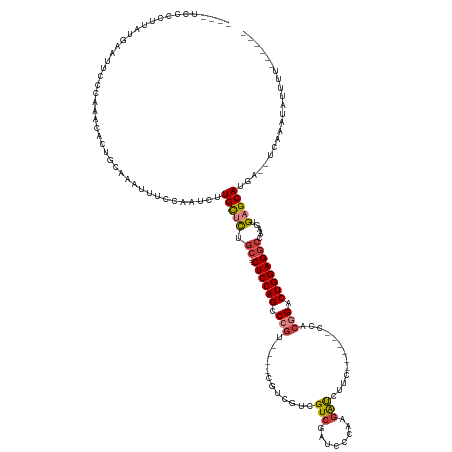
| Sequence ID | dm3.chrX |
|---|---|
| Location | 21,004,521 – 21,004,643 |
| Length | 122 |
| Max. P | 0.989860 |

| Location | 21,004,521 – 21,004,643 |
|---|---|
| Length | 122 |
| Sequences | 7 |
| Columns | 147 |
| Reading direction | forward |
| Mean pairwise identity | 70.10 |
| Shannon entropy | 0.54291 |
| G+C content | 0.48160 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -16.81 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21004521 122 + 22422827 ----UCCUCUUAUAAAUUCCCAAAUACUGCAAAUUUCCAACCUUGCUCUGC---CUCCAGCCCGU----CGUCGUCGUCGAUACGAAGAUCUUC------CCACGGACUGGAGGCCAAGUGAGCAUGA--CCAAAUAUUUU------ ----.................................((..((..((..((---((((((.((((----.(((.((((....)))).)))....------..)))).))))))))..))..))..)).--...........------ ( -31.70, z-score = -3.15, R) >droSim1.chrX 16145524 122 + 17042790 ----UCCCCUUAUAAAUUCCCAAGCACUGCAAAUUUCCAAUCUUGCUCUGC---CUCCAGCCCGC----CGUCGUCGUCGAUCCCAAGAUCUUC------CCACGGACUGGAGGCCAAGUGAGCAUGA--UCAAAUUUUUU------ ----..........................((((((...(((.(((((.((---((((((.(((.----((....))..((((....))))...------...))).)))))))).....))))).))--).))))))...------ ( -31.90, z-score = -2.97, R) >droSec1.super_8 3288300 122 + 3762037 ----UCCCCUUAUAAAUUCCCAAACACUGCAAAUUUCCAAUCUUGCUCUGC---CUCCAGCCCGU----CGUCGUCGUCGAUCCCAAGAUCUUC------CCACGGACUGGAGGCCAAGUGAGCAUGA--UCAAAUAUUUU------ ----...................................(((.(((((.((---((((((.((((----((....))..((((....))))...------..)))).)))))))).....))))).))--)..........------ ( -32.30, z-score = -3.80, R) >droYak2.chrX 20204524 122 + 21770863 ----UACCCUUAUGAAUUCCCAAAAACUGCAAAUUGCCAACAUUGCUCUGC---CUCCAGACCGU----CGUCGUCGUCGAUCCGCAGACCUUC------CCACGGACUGGAGGCCAAGUGAGCAUGA--CGAAAUAUUUU------ ----............(((.........((.....))...(((.((((.((---((((((.((((----.(..(..(((........)))..).------.))))).)))))))).....))))))).--.))).......------ ( -31.80, z-score = -2.14, R) >droEre2.scaffold_4690 17569800 123 + 18748788 -----UCCCCUAUGAAUUCCCAAAAACUGCAAAUUGCCAAUGUUGCUCUGC---CUCCAGACCGU----CGUCGUCGUCGAUCCGCAGAUCUUC------CCACGGACUGGAGGCCAAGUGAGCAUAAACUCUAAUAUUUU------ -----.......................((.....))......(((((.((---((((((.((((----((....))..((((....))))...------..)))).)))))))).....)))))................------ ( -30.30, z-score = -2.12, R) >droAna3.scaffold_13334 347540 132 - 1562580 --------UAUAUGUAUUAAU-UGCACUGUUACUUACUGUUACUGUGUCCAGACCUCCAGUCCUCAGAGCGCACUCGUCGAUCCCCAGAUUUAG------ACACAGACUGGAGGCCAAGUGAGCAUACUUUCAUCCAUAUUUUAUCU --------..(((((......-.((((.((.((.....)).)).))))...(.(((((((((....(((....)))(((((((....))))..)------))...))))))))).)......))))).................... ( -31.60, z-score = -1.69, R) >droWil1.scaffold_180702 10189 134 + 4511350 UUAGAGACAUAGAGAGAUUGAGUGCAAUCCAUAUUACUGA-GCUGCCUUGCGUCCUCCAGGCGGU----CGUCGUCGUCGUCGUCGAAGCCUUAAUCAAACCUCAGACUGGAGGCCAUGUGAGCACUA--UGAACUAUUGU------ .......(((((.(.(((((....))))))..........-..(((.(..((((((((((..(((----..(((.((....)).))).)))....((........)))))))))..)))..)))))))--)).........------ ( -35.20, z-score = 0.45, R) >consensus ____UCCCCUUAUGAAUUCCCAAACACUGCAAAUUUCCAAUCUUGCUCUGC___CUCCAGCCCGU____CGUCGUCGUCGAUCCCAAGAUCUUC______CCACGGACUGGAGGCCAAGUGAGCAUGA__UCAAAUAUUUU______ ...........................................(((((......((((((.(((............(((........))).............))).)))))).......)))))...................... (-16.81 = -17.02 + 0.21)


| Location | 21,004,521 – 21,004,643 |
|---|---|
| Length | 122 |
| Sequences | 7 |
| Columns | 147 |
| Reading direction | reverse |
| Mean pairwise identity | 70.10 |
| Shannon entropy | 0.54291 |
| G+C content | 0.48160 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -18.34 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 21004521 122 - 22422827 ------AAAAUAUUUGG--UCAUGCUCACUUGGCCUCCAGUCCGUGG------GAAGAUCUUCGUAUCGACGACGACG----ACGGGCUGGAG---GCAGAGCAAGGUUGGAAAUUUGCAGUAUUUGGGAAUUUAUAAGAGGA---- ------.......((..--((.(((((.....(((((((((((((.(------.....((.((((....)))).))).----)))))))))))---)).))))).))..))((((((.(((...))).)))))).........---- ( -43.30, z-score = -3.16, R) >droSim1.chrX 16145524 122 - 17042790 ------AAAAAAUUUGA--UCAUGCUCACUUGGCCUCCAGUCCGUGG------GAAGAUCUUGGGAUCGACGACGACG----GCGGGCUGGAG---GCAGAGCAAGAUUGGAAAUUUGCAGUGCUUGGGAAUUUAUAAGGGGA---- ------.......(..(--((.(((((.....(((((((((((((.(------...((((....))))........).----)))))))))))---)).))))).)))..)((((((.(((...))).)))))).........---- ( -47.20, z-score = -3.89, R) >droSec1.super_8 3288300 122 - 3762037 ------AAAAUAUUUGA--UCAUGCUCACUUGGCCUCCAGUCCGUGG------GAAGAUCUUGGGAUCGACGACGACG----ACGGGCUGGAG---GCAGAGCAAGAUUGGAAAUUUGCAGUGUUUGGGAAUUUAUAAGGGGA---- ------.......(..(--((.(((((.....(((((((((((((.(------...((((....))))........).----)))))))))))---)).))))).)))..)((((((.(((...))).)))))).........---- ( -46.90, z-score = -4.35, R) >droYak2.chrX 20204524 122 - 21770863 ------AAAAUAUUUCG--UCAUGCUCACUUGGCCUCCAGUCCGUGG------GAAGGUCUGCGGAUCGACGACGACG----ACGGUCUGGAG---GCAGAGCAAUGUUGGCAAUUUGCAGUUUUUGGGAAUUCAUAAGGGUA---- ------..........(--((((((((.....((((((((.((((.(------....(((..((...))..)))..).----)))).))))))---)).)))))....))))(((((.(((...))).)))))..........---- ( -39.40, z-score = -1.14, R) >droEre2.scaffold_4690 17569800 123 - 18748788 ------AAAAUAUUAGAGUUUAUGCUCACUUGGCCUCCAGUCCGUGG------GAAGAUCUGCGGAUCGACGACGACG----ACGGUCUGGAG---GCAGAGCAACAUUGGCAAUUUGCAGUUUUUGGGAAUUCAUAGGGGA----- ------.........((((((.(((((.....((((((((.((((.(------...((((....))))........).----)))).))))))---)).))))).((..(((........)))..)).))))))........----- ( -41.70, z-score = -2.44, R) >droAna3.scaffold_13334 347540 132 + 1562580 AGAUAAAAUAUGGAUGAAAGUAUGCUCACUUGGCCUCCAGUCUGUGU------CUAAAUCUGGGGAUCGACGAGUGCGCUCUGAGGACUGGAGGUCUGGACACAGUAACAGUAAGUAACAGUGCA-AUUAAUACAUAUA-------- .......(((((.((....)).(((.(.((.((((((((((((..((------(...(((....))).)))(((....)))...)))))))))))).)).....((.((.....)).)).).)))-.......))))).-------- ( -36.40, z-score = -0.96, R) >droWil1.scaffold_180702 10189 134 - 4511350 ------ACAAUAGUUCA--UAGUGCUCACAUGGCCUCCAGUCUGAGGUUUGAUUAAGGCUUCGACGACGACGACGACG----ACCGCCUGGAGGACGCAAGGCAGC-UCAGUAAUAUGGAUUGCACUCAAUCUCUCUAUGUCUCUAA ------....(((..((--(((.(((..(.((((((((((.(((((((((.....)))))))).((.((....)).))----...).))))))).).)).)..)))-..........((((((....))))))..)))))...))). ( -34.00, z-score = 0.73, R) >consensus ______AAAAUAUUUGA__UCAUGCUCACUUGGCCUCCAGUCCGUGG______GAAGAUCUGGGGAUCGACGACGACG____ACGGGCUGGAG___GCAGAGCAAGAUUGGAAAUUUGCAGUGUUUGGGAAUUCAUAAGGGGA____ ......................(((((.......((((((.((((...........((((....))))..............)))).))))))......)))))........................................... (-18.34 = -19.20 + 0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:47 2011