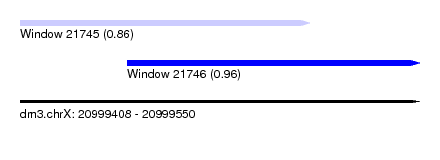
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,999,408 – 20,999,550 |
| Length | 142 |
| Max. P | 0.964207 |

| Location | 20,999,408 – 20,999,511 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.51 |
| Shannon entropy | 0.35028 |
| G+C content | 0.47656 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -19.92 |
| Energy contribution | -19.72 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
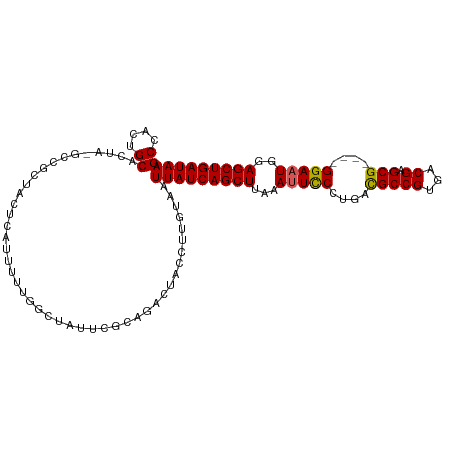
>dm3.chrX 20999408 103 + 22422827 GCCACUGCACUA-GCCGCUACUCAUUUUUAGCUAUUCGCAGACUACCUUGUAAUUAUCAGCUUAGAUUCCCUGACGCCGUGACGAGCG----GGAAUGGAGCUGAUAA ....((((..((-((...............))))...))))............((((((((((..((((((...((......))...)----))))).)))))))))) ( -28.36, z-score = -1.59, R) >droEre2.scaffold_4690 17565409 96 + 18748788 GCGAAAGCAAUGCAUCAUUUUUCCUAUUCGG-------C-GACUACCUUGCAAUUAUCAGCUCAAAUUCCCUACUGCCGUGGCGAGCG----GGAAUGCAGCUGAUAA ((....))..((((........((.....))-------.-........)))).(((((((((...((((((...(((....)))...)----)))))..))))))))) ( -31.43, z-score = -2.58, R) >droYak2.chrX 20200709 103 + 21770863 GCCACUGCAAUGCAUCAUUUUUCCUAUUCGGAGACUAAC-GACUACCUUGUAAUUAUCAGCUUAAAUUUCCUACUGCCGCGACGAGCG----GGAAUGUAGCUGAUAA .....(((((.........(((((.....)))))(....-)......))))).(((((((((...((((((....((((...)).)))----)))))..))))))))) ( -22.90, z-score = -1.23, R) >droSec1.super_8 3283087 107 + 3762037 GCCACUGCACUA-GCCGCUACUCAUUUUUUGCUAUUCGCAGACUACCUUGUAAUUAUCAGCUUAAAGUCCCUGACGCCGUGACGAGCGAGCGGGAAUGGAGCUGAUAA ....((((..((-((...............))))...))))............((((((((((....((((...(((((...)).)))...))))...)))))))))) ( -30.46, z-score = -1.38, R) >droSim1.chrX 861641 107 - 17042790 GCCACUGCACUA-GCCGCUACUCAUUUUUUGCUAUUCGCAGACUACCUUGUAAUUAUCAGCUUAAAUUCCCUGACGCCGUGACGAGCGAGCGGGAAUGGAGCUGAUAA ....((((..((-((...............))))...))))............((((((((((..((((((...(((((...)).)))...)))))).)))))))))) ( -31.16, z-score = -2.12, R) >consensus GCCACUGCACUA_GCCGCUACUCAUUUUUGGCUAUUCGCAGACUACCUUGUAAUUAUCAGCUUAAAUUCCCUGACGCCGUGACGAGCG____GGAAUGGAGCUGAUAA ((....)).............................................(((((((((...(((((....(((((...)).)))....)))))..))))))))) (-19.92 = -19.72 + -0.20)

| Location | 20,999,446 – 20,999,550 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Shannon entropy | 0.34288 |
| G+C content | 0.47582 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.16 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20999446 104 + 22422827 -----AGACUACCUUGUAAUUAUCAGCUUAGAUUCCCUGACGCCGUGACGAGCG----GGAAUGGAGCUGAUAAGGCGCUCGCAAAUAACCAUUUUACACCAUGUACGACUAU -----.........(((((((((((((((..((((((...((......))...)----))))).)))))))))).((....))...........))))).............. ( -26.00, z-score = -1.55, R) >droEre2.scaffold_4690 17565440 86 + 18748788 -----CGACUACCUUGCAAUUAUCAGCUCAAAUUCCCUACUGCCGUGGCGAGCG----GGAAUGCAGCUGAUAAGACGCUCGCCAGUAACCAUGC------------------ -----..(((..(..((..(((((((((...((((((...(((....)))...)----)))))..)))))))))...))..)..)))........------------------ ( -27.20, z-score = -2.08, R) >droYak2.chrX 20200742 98 + 21770863 ACUAACGACUACCUUGUAAUUAUCAGCUUAAAUUUCCUACUGCCGCGACGAGCG----GGAAUGUAGCUGAUAAGCCGCUCGCAAAUAACCAUUU-ACACCAA---------- ..............((((((((((((((...((((((....((((...)).)))----)))))..)))))))))((.....))..........))-)))....---------- ( -21.20, z-score = -1.76, R) >droSec1.super_8 3283125 107 + 3762037 -----AGACUACCUUGUAAUUAUCAGCUUAAAGUCCCUGACGCCGUGACGAGCGAGCGGGAAUGGAGCUGAUAAGCCGCUCGCAAAUAACCAUUU-ACACCAUCUACGACUAU -----.........(((((((((((((((....((((...(((((...)).)))...))))...))))))))))((.....))..........))-))).............. ( -27.40, z-score = -1.84, R) >droSim1.chrX 861679 107 - 17042790 -----AGACUACCUUGUAAUUAUCAGCUUAAAUUCCCUGACGCCGUGACGAGCGAGCGGGAAUGGAGCUGAUAAGCCGCUCGCAAAUAACCAUUU-ACACCAUCUACGACUAU -----.........(((((((((((((((..((((((...(((((...)).)))...)))))).))))))))))((.....))..........))-))).............. ( -28.10, z-score = -2.56, R) >consensus _____AGACUACCUUGUAAUUAUCAGCUUAAAUUCCCUGACGCCGUGACGAGCG____GGAAUGGAGCUGAUAAGCCGCUCGCAAAUAACCAUUU_ACACCAU_UACGACUAU ...............((..(((((((((...(((((....(((((...)).)))....)))))..)))))))))...)).................................. (-20.52 = -20.16 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:45 2011