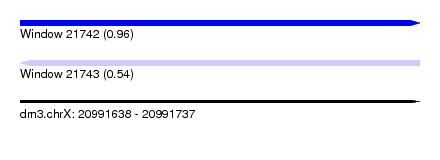
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,991,638 – 20,991,737 |
| Length | 99 |
| Max. P | 0.956758 |

| Location | 20,991,638 – 20,991,737 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Shannon entropy | 0.41772 |
| G+C content | 0.56322 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -26.68 |
| Energy contribution | -28.03 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
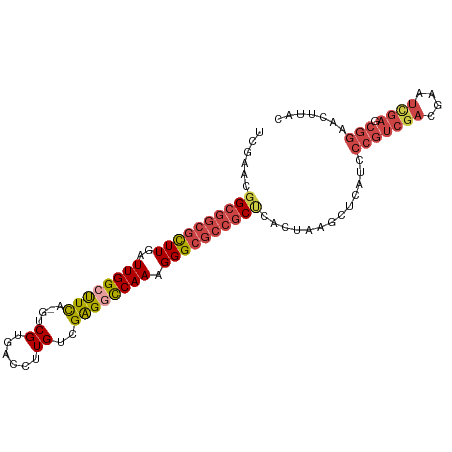
>dm3.chrX 20991638 99 + 22422827 UCGGACGGCGGCGCUUGAUUGGGUUCA-GUCGUGACCUUGUCGAGGCCAAAGGGCGCCGCUCACUAAGCUCAUCCCGUCGACGAAUUGAGCGGAACUUAC ......((((((.((((((.(((((..-.....))))).))))))(((....)))))))))......(((((...((....))...)))))......... ( -36.10, z-score = -0.58, R) >droSim1.chrX_random 258799 99 - 5698898 UCGAACGGCGGCGCUUGAUUGGCUUCA-GUCGUGACCUUGUCGAGGCCAAAGGGCGCCGCCCACUAAGCUCAUCCCGUCGACGAAUCGAGCGGAACUUCC ......((((((((((..((((((((.-(.((......)).))))))))).)))))))))).............(((((((....)))).)))....... ( -41.20, z-score = -1.97, R) >droSec1.super_8 3274907 99 + 3762037 UCGAACGGCGGCGCUUGAUUGGCUUCA-GUCGUAACCUUGUCGAGGCCAAAGGGCGCCGCCCACUAAGCUCAUCCCGUCGACGAAUCGAGCGGAACUUAC ......((((((((((..((((((((.-(.((......)).))))))))).)))))))))).............(((((((....)))).)))....... ( -41.20, z-score = -2.63, R) >droYak2.chrX 20192087 99 + 21770863 UCGAACGGCGGCGCUUGAUUGGCUUCA-CUCGUGACCUUGUUGAGGCCAAAGGGCGCCGCUUACUAAGCUCAUCCCGUCGACGAAUCGAGCGGAACUUAC ......((((((((((..(((((((((-..((......)).))))))))).)))))))))).............(((((((....)))).)))....... ( -39.50, z-score = -2.62, R) >droEre2.scaffold_4690 17557935 99 + 18748788 UAAAACGGCGGCGUUUGAUUGGGCUCA-GUCGUGACCUUGCUGAGGCCAAAGGGCGCCGCUCACUAAGCUCAUCCCGUCGACUAAUUGAGCGGAACUUAU ......((((((((((..((((.((((-(.((......))))))).)))).))))))))))......(((((..............)))))......... ( -37.84, z-score = -2.25, R) >droAna3.scaffold_13334 333147 80 - 1562580 CUGAAAAGCGGCGCUUGAUUGGCUUUAAGGCGUGACCUUGUGGGGGUCAAAGGGGCAAUCGUAAAAAUUACAAAAAUUCU-------------------- ..(((((((...))))(((((.((((......((((((.....)))))).)))).)))))................))).-------------------- ( -17.30, z-score = -0.04, R) >consensus UCGAACGGCGGCGCUUGAUUGGCUUCA_GUCGUGACCUUGUCGAGGCCAAAGGGCGCCGCUCACUAAGCUCAUCCCGUCGACGAAUCGAGCGGAACUUAC ......((((((((((..((((((((....((......))..)))))))).)))))))))).............(((((((....)))).)))....... (-26.68 = -28.03 + 1.35)


| Location | 20,991,638 – 20,991,737 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Shannon entropy | 0.41772 |
| G+C content | 0.56322 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -21.15 |
| Energy contribution | -24.48 |
| Covariance contribution | 3.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
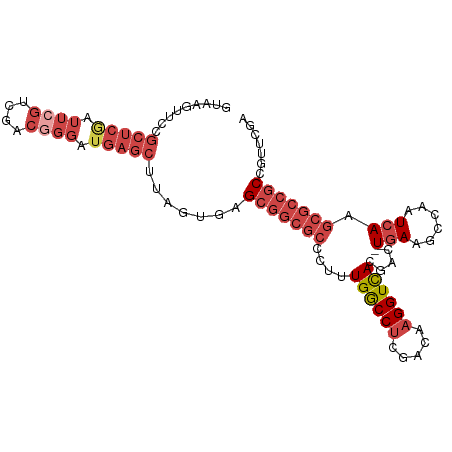
>dm3.chrX 20991638 99 - 22422827 GUAAGUUCCGCUCAAUUCGUCGACGGGAUGAGCUUAGUGAGCGGCGCCCUUUGGCCUCGACAAGGUCACGAC-UGAACCCAAUCAAGCGCCGCCGUCCGA .........(((((.((((....)))).))))).....(.(((((((....((((((.....)))))).((.-((....)).))..))))))))...... ( -33.40, z-score = -0.64, R) >droSim1.chrX_random 258799 99 + 5698898 GGAAGUUCCGCUCGAUUCGUCGACGGGAUGAGCUUAGUGGGCGGCGCCCUUUGGCCUCGACAAGGUCACGAC-UGAAGCCAAUCAAGCGCCGCCGUUCGA ....((((((.((((....))))))))))((((......((((((((...(((((.(((....(....)...-))).)))))....)))))))))))).. ( -39.60, z-score = -0.83, R) >droSec1.super_8 3274907 99 - 3762037 GUAAGUUCCGCUCGAUUCGUCGACGGGAUGAGCUUAGUGGGCGGCGCCCUUUGGCCUCGACAAGGUUACGAC-UGAAGCCAAUCAAGCGCCGCCGUUCGA ((..((((((.((((....))))))))))..))......((((((((...(((((.(((....(....)...-))).)))))....))))))))...... ( -40.00, z-score = -1.56, R) >droYak2.chrX 20192087 99 - 21770863 GUAAGUUCCGCUCGAUUCGUCGACGGGAUGAGCUUAGUAAGCGGCGCCCUUUGGCCUCAACAAGGUCACGAG-UGAAGCCAAUCAAGCGCCGCCGUUCGA ((..((((((.((((....))))))))))..)).......(((((((...(((((.(((....(....)...-))).)))))....)))))))....... ( -37.80, z-score = -2.04, R) >droEre2.scaffold_4690 17557935 99 - 18748788 AUAAGUUCCGCUCAAUUAGUCGACGGGAUGAGCUUAGUGAGCGGCGCCCUUUGGCCUCAGCAAGGUCACGAC-UGAGCCCAAUCAAACGCCGCCGUUUUA .(((((((..(((...........)))..)))))))..(.((((((....((((.(((((...(....)..)-)))).)))).....)))))))...... ( -31.30, z-score = -0.88, R) >droAna3.scaffold_13334 333147 80 + 1562580 --------------------AGAAUUUUUGUAAUUUUUACGAUUGCCCCUUUGACCCCCACAAGGUCACGCCUUAAAGCCAAUCAAGCGCCGCUUUUCAG --------------------.(((................(((((......(((((.......))))).((......)))))))((((...))))))).. ( -11.50, z-score = -0.74, R) >consensus GUAAGUUCCGCUCGAUUCGUCGACGGGAUGAGCUUAGUGAGCGGCGCCCUUUGGCCUCGACAAGGUCACGAC_UGAAGCCAAUCAAGCGCCGCCGUUCGA .........(((((.((((....)))).))))).......(((((((...(((((.(((....(....)....))).)))))....)))))))....... (-21.15 = -24.48 + 3.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:42 2011