
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,966,740 – 20,966,896 |
| Length | 156 |
| Max. P | 0.848152 |

| Location | 20,966,740 – 20,966,896 |
|---|---|
| Length | 156 |
| Sequences | 6 |
| Columns | 162 |
| Reading direction | reverse |
| Mean pairwise identity | 63.08 |
| Shannon entropy | 0.70664 |
| G+C content | 0.48154 |
| Mean single sequence MFE | -42.89 |
| Consensus MFE | -20.81 |
| Energy contribution | -22.52 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20966740 156 - 22422827 GGUCACCACAAUUGGCCACACAACUGCAGCAAGUGUGGCCAGGUCUUUCGCACAUCAGGCUUUAUGCGCAUGCACUCCGUGAAUCCCUGUGAGCGAAACUUACACAAGUUUAAAAUAACUAA--GCCAAAUGAAUUGCAGCAACUUG----GCAAAAAACCA (((........((((((((((...........))))))))))...((((((.((.((((.((((((.(........)))))))..)))))).))))))........((((......))))..--(((((.((........))..)))----)).....))). ( -40.70, z-score = -0.41, R) >droSim1.chrX_random 5404806 153 - 5698898 GGUCACCACAAUUGGCCAUACAACUGCAGCAAGUGUGGCCAGGUCUUCCGUACAUCAGGCUUUAUGCGCAUGCACUCCGUGAAUGCCUGUGAGCGCAACUUGCACAAGUUUAAAAUAUCUAA--GCCAAAUUAAUU---GCAACUUG----GCCAAACCGCA (((........(((((((......((((((((((...((..((....))((.((.(((((((((((.(........))))))).))))))).)))).))))))....(((((.......)))--)).........)---)))...))----))))))))... ( -42.30, z-score = -0.03, R) >droSec1.super_8 3253760 156 - 3762037 GGUCACCACAAUUGGCCACACAACUGCAGCAAGUGUGGCCAGGUCUUCCGUACAUCAGGCUUUAUGCGCAUGCACUCCGUGAAUGCCUGUGAGCGCAACUUGCACAAGUUUAAAAUAUCUAA--GCCAAAUUAAUUGCAGCAACUUG----GCCAAACCGCA (((........(((((((.....(((((((((((...((..((....))((.((.(((((((((((.(........))))))).))))))).)))).))))))....(((((.......)))--)).........))))).....))----))))))))... ( -45.80, z-score = -0.43, R) >droYak2.chr2R 19607212 136 + 21139217 ------GAGCACUAAAAAGAAGUCUACAGAGAGGGUAAACACGUUUUUUGCCGAGGAACCAUUGGUAGAAUGAGAGAAGUGAGUGGUUCCAAAACAAAACAGCAACUGAGCGGACUGAAAAAACGCUAAGCAAACAAGAGCA-------------------- ------.(((.((....)).(((((.(((....((((((.......))))))..(((((((((.(..............).)))))))))...............)))...)))))........)))..((........)).-------------------- ( -27.04, z-score = -0.17, R) >droEre2.scaffold_4690 17536605 147 - 18748788 GGCCACCACACCUGGUCACACAAUUGCGGCAAGUGUGGCCAGGUCUUCCGCACAGAAGGCUUCAAGCGCAUGCACUCCUCUAAUGCCUGUGAGCGCAAUCUGAGCAAGUUCAAAAUCGCUAA--GCCGAAUGAACCUAAGCCAAGAACA------------- (((......((((((((((((...........)))))))))))).....((.((((..((((((((.((((...........)))))).)))).))..)))).))..(((((...(((....--..))).)))))....))).......------------- ( -46.90, z-score = -1.35, R) >droAna3.scaffold_13334 311966 158 + 1562580 GGCCACCACAACUGGUCACACAAGUGCGAGAAGUGUGGCCAGAUCUUUCGCACGGCAGGCUUCAAGAGGAUGCAUGCCGAGGGAGCCUGUGAGCGCAAUUUGCUGAAACAGAAACUGA--AA--ACCAAAUGAAAUUAAGCAACUUGUAUAGCGGGGGAACA ((((((.((..((.(.(((....)))).))..)))))))).(.(((..(((...(((((((((....((.......))...)))))))))....((((.(((((...........((.--..--..))..........))))).))))...)))..))).). ( -54.60, z-score = -2.44, R) >consensus GGUCACCACAACUGGCCACACAACUGCAGCAAGUGUGGCCAGGUCUUCCGCACAGCAGGCUUUAUGCGCAUGCACUCCGUGAAUGCCUGUGAGCGCAACUUGCACAAGUUUAAAAUAACUAA__GCCAAAUGAAUUGAAGCAACUUG____GC_AAA___CA (((........((((((((((...........))))))))))......(((.((.(((((((((((...........)))))).))))))).))).............................)))................................... (-20.81 = -22.52 + 1.70)

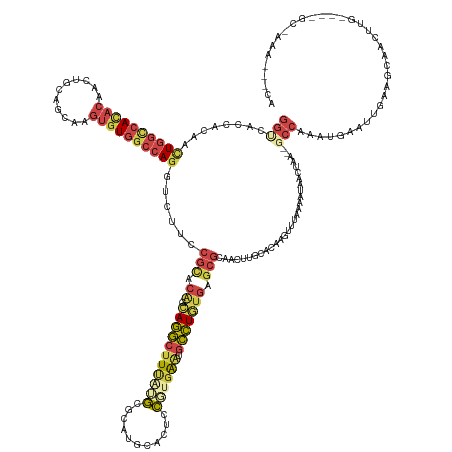
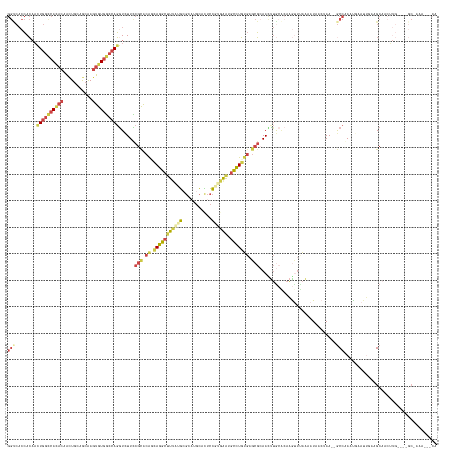
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:38 2011