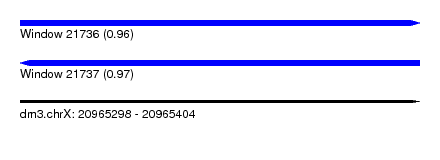
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,965,298 – 20,965,404 |
| Length | 106 |
| Max. P | 0.971858 |

| Location | 20,965,298 – 20,965,404 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 86.38 |
| Shannon entropy | 0.23961 |
| G+C content | 0.54212 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -26.24 |
| Energy contribution | -27.68 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20965298 106 + 22422827 ---GCAGCAGCAUCAGCAAGCCGGUGUAAUCAACACGAUCUCUGGCAGCAUUCCGGCCACGGGAUUUAGUUCCCUGGCCGGCAGCUUUAGUGAUCUUCAGUUGAACAAC ---((....)).(((((......((((.....))))((((.((((.(((...(((((((.((((......)))))))))))..))))))).))))....)))))..... ( -42.40, z-score = -4.09, R) >droSim1.chrX 16123211 106 + 17042790 ---GCAGCAGCAUCAGCAAGCUGGCCUAAUCAACACGAUCUCUGGCAGCAUUCCGGCCACGGGAUUUAGUUCCCUGGCCGGCAGCUUUAGUGAUCUUCAGUUGAACAAC ---.((((.((....))..))))......(((((..((((.((((.(((...(((((((.((((......)))))))))))..))))))).))))....)))))..... ( -42.50, z-score = -3.96, R) >droSec1.super_8 3252343 106 + 3762037 ---GCAGCAGCAUCAGCAAGCUGGCCUAAUCAGCACGAUCUCUGGCAGCAUUCCGGCCACGGGAUUUAGUUCCCUGGCCGGCAGCUUUAGUGAUCUUCAGUUGAACAAC ---((....)).(((((..(((((.....)))))..((((.((((.(((...(((((((.((((......)))))))))))..))))))).))))....)))))..... ( -44.80, z-score = -4.12, R) >droYak2.chrX 20168670 109 + 21770863 GCAGCAGCAGCAUCAGCAAACUGGCCUGAUCAACACGAUCUCCGGCAGCAUUCCGGCCACGGGAUUCAGUUCUCUGGCCGGCAGCUUCAGUGAUCUGCAUUUGAACAAC ......((((.(((.....(((((((.((((.....))))...)))(((...(((((((.((((......)))))))))))..))).)))))))))))........... ( -39.40, z-score = -2.34, R) >droVir3.scaffold_12970 7717273 106 + 11907090 ---GCAGCAACAUCUGCAGACUGGCGUCAUCAACACCAUUUCCGGCAGCAUUCCAGCAACUGGCUUUACAUCGCUGGCGGGCAGCUUUAGCGACUUGCAGCUAAACAAU ---..........((((....(((.((.....)).)))...(((.((((.....(((.....))).......)))).))))))).((((((........)))))).... ( -27.00, z-score = 0.46, R) >consensus ___GCAGCAGCAUCAGCAAGCUGGCCUAAUCAACACGAUCUCUGGCAGCAUUCCGGCCACGGGAUUUAGUUCCCUGGCCGGCAGCUUUAGUGAUCUUCAGUUGAACAAC ....((((.......((......))...........((((.((((.(((...(((((((.((((......)))))))))))..))))))).))))....))))...... (-26.24 = -27.68 + 1.44)

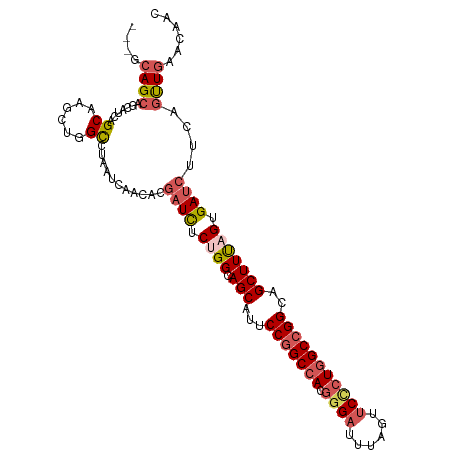
| Location | 20,965,298 – 20,965,404 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.38 |
| Shannon entropy | 0.23961 |
| G+C content | 0.54212 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -32.98 |
| Energy contribution | -34.18 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20965298 106 - 22422827 GUUGUUCAACUGAAGAUCACUAAAGCUGCCGGCCAGGGAACUAAAUCCCGUGGCCGGAAUGCUGCCAGAGAUCGUGUUGAUUACACCGGCUUGCUGAUGCUGCUGC--- ((...(((((....((((.((..(((..(((((((((((......)))).)))))))...)))...)).))))..)))))..))..((((..((....)).)))).--- ( -44.20, z-score = -4.28, R) >droSim1.chrX 16123211 106 - 17042790 GUUGUUCAACUGAAGAUCACUAAAGCUGCCGGCCAGGGAACUAAAUCCCGUGGCCGGAAUGCUGCCAGAGAUCGUGUUGAUUAGGCCAGCUUGCUGAUGCUGCUGC--- (((..(((((....((((.((..(((..(((((((((((......)))).)))))))...)))...)).))))..)))))...)))((((..((....)).)))).--- ( -45.20, z-score = -4.04, R) >droSec1.super_8 3252343 106 - 3762037 GUUGUUCAACUGAAGAUCACUAAAGCUGCCGGCCAGGGAACUAAAUCCCGUGGCCGGAAUGCUGCCAGAGAUCGUGCUGAUUAGGCCAGCUUGCUGAUGCUGCUGC--- (((..(((.(....((((.((..(((..(((((((((((......)))).)))))))...)))...)).))))..).)))...)))((((..((....)).)))).--- ( -41.00, z-score = -2.53, R) >droYak2.chrX 20168670 109 - 21770863 GUUGUUCAAAUGCAGAUCACUGAAGCUGCCGGCCAGAGAACUGAAUCCCGUGGCCGGAAUGCUGCCGGAGAUCGUGUUGAUCAGGCCAGUUUGCUGAUGCUGCUGCUGC ..((.((((.(((.((((.(((.(((..((((((((.((......)).).)))))))...)))..))).))))))))))).))(((.(((..((....)).)))))).. ( -37.80, z-score = -0.91, R) >droVir3.scaffold_12970 7717273 106 - 11907090 AUUGUUUAGCUGCAAGUCGCUAAAGCUGCCCGCCAGCGAUGUAAAGCCAGUUGCUGGAAUGCUGCCGGAAAUGGUGUUGAUGACGCCAGUCUGCAGAUGUUGCUGC--- ........((.(((((((......((.((...((((((((.........))))))))...)).))((((..(((((((...))))))).))))..))).)))).))--- ( -33.40, z-score = 0.00, R) >consensus GUUGUUCAACUGAAGAUCACUAAAGCUGCCGGCCAGGGAACUAAAUCCCGUGGCCGGAAUGCUGCCAGAGAUCGUGUUGAUUAGGCCAGCUUGCUGAUGCUGCUGC___ (((..(((((....((((.((..(((..(((((((((((......)))).)))))))...)))...)).))))..)))))...)))((((..((....)).)))).... (-32.98 = -34.18 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:37 2011