| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,225,728 – 12,225,791 |
| Length | 63 |
| Max. P | 0.979033 |

| Location | 12,225,728 – 12,225,791 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 67 |
| Reading direction | forward |
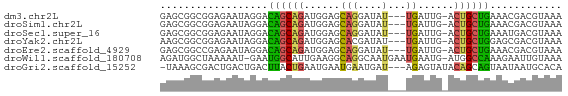
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.61814 |
| G+C content | 0.45411 |
| Mean single sequence MFE | -7.33 |
| Consensus MFE | -5.63 |
| Energy contribution | -5.08 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.83 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979033 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
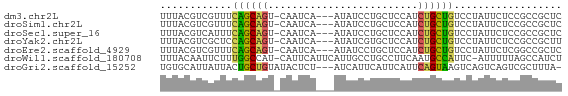
>dm3.chr2L 12225728 63 + 23011544 UUUACGUCGUUUCAGCAGU-CAAUCA---AUAUCCUGCUCCAUCUGCUGUCCUAUUCUCCGCCGCUC ............((((((.-....((---......))......)))))).................. ( -7.30, z-score = -1.50, R) >droSim1.chr2L 12022203 63 + 22036055 UUUACGUCGUUUCAGCAGU-CAAUCA---AUAUCCUGCUCCAUCUGCUGUCCUAUUCUCCGCCGCUC ............((((((.-....((---......))......)))))).................. ( -7.30, z-score = -1.50, R) >droSec1.super_16 416228 63 + 1878335 UUUACGUCAUUUCAGCAGU-CAAUCA---AUAUCCUGCUCCAUCUGCUGUCCUAUUCUCCGCCGCUC ............((((((.-....((---......))......)))))).................. ( -7.30, z-score = -1.82, R) >droYak2.chr2L 8651498 63 + 22324452 UUUACGUCGCUCCAGCAGU-CAAUCA---AUAUCGUGCUCCAUCUGCUGUCCUAUUCUCCGCCGCUU ............((((((.-....((---......))......)))))).................. ( -7.30, z-score = -0.51, R) >droEre2.scaffold_4929 13449436 63 - 26641161 UUUACGUCGUUUCAGCAGU-CAAUCA---AUAUCCUGCUCCAUCUGCUGUCCUAUUCUCGGCCGCUC .....((((...((((((.-....((---......))......)))))).........))))..... ( -9.80, z-score = -1.74, R) >droWil1.scaffold_180708 2657312 65 - 12563649 UUUACAAUUCUUUGGCCAU-CAUUCAUUCAUUGCCUGCCUUCAAUGCCAUUC-AUUUUUAGCCAUCU ............((((...-........(((((........)))))......-.......))))... ( -5.75, z-score = 0.04, R) >droGri2.scaffold_15252 14717034 63 + 17193109 UGUGCAUUAUUACUGCUGUAUACUCU---AUCAUUCAUUCAUUCAGUAAGUCAGUCAGUCGCUUUA- .(((.......(((((((..((((..---...............))))...))).)))))))....- ( -6.54, z-score = 0.11, R) >consensus UUUACGUCGUUUCAGCAGU_CAAUCA___AUAUCCUGCUCCAUCUGCUGUCCUAUUCUCCGCCGCUC ............((((((.........................)))))).................. ( -5.63 = -5.08 + -0.55)
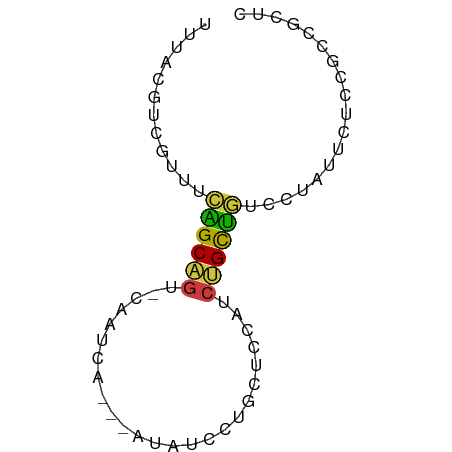
| Location | 12,225,728 – 12,225,791 |
|---|---|
| Length | 63 |
| Sequences | 7 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.61814 |
| G+C content | 0.45411 |
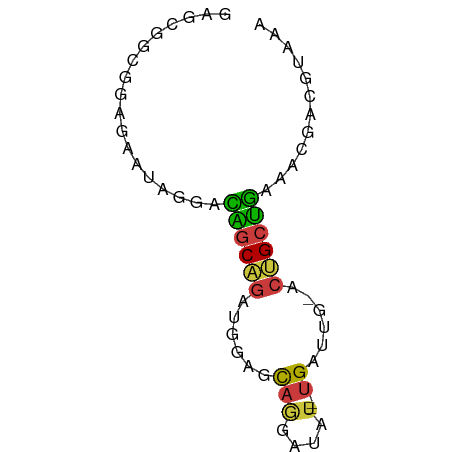
| Mean single sequence MFE | -11.72 |
| Consensus MFE | -5.76 |
| Energy contribution | -5.23 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936471 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
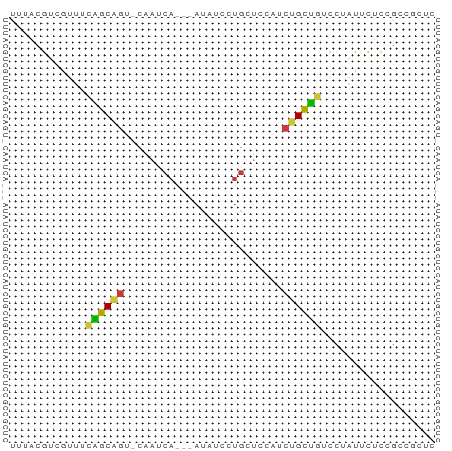
>dm3.chr2L 12225728 63 - 23011544 GAGCGGCGGAGAAUAGGACAGCAGAUGGAGCAGGAUAU---UGAUUG-ACUGCUGAAACGACGUAAA ..(((.((..........((((((.(.((.(((....)---)).)).-)))))))...)).)))... ( -11.52, z-score = -1.39, R) >droSim1.chr2L 12022203 63 - 22036055 GAGCGGCGGAGAAUAGGACAGCAGAUGGAGCAGGAUAU---UGAUUG-ACUGCUGAAACGACGUAAA ..(((.((..........((((((.(.((.(((....)---)).)).-)))))))...)).)))... ( -11.52, z-score = -1.39, R) >droSec1.super_16 416228 63 - 1878335 GAGCGGCGGAGAAUAGGACAGCAGAUGGAGCAGGAUAU---UGAUUG-ACUGCUGAAAUGACGUAAA ..(((.(........)..((((((.(.((.(((....)---)).)).-)))))))......)))... ( -11.10, z-score = -1.34, R) >droYak2.chr2L 8651498 63 - 22324452 AAGCGGCGGAGAAUAGGACAGCAGAUGGAGCACGAUAU---UGAUUG-ACUGCUGGAGCGACGUAAA .....(((..(.......((((((.(.((.((......---)).)).-)))))))...)..)))... ( -12.60, z-score = -0.97, R) >droEre2.scaffold_4929 13449436 63 + 26641161 GAGCGGCCGAGAAUAGGACAGCAGAUGGAGCAGGAUAU---UGAUUG-ACUGCUGAAACGACGUAAA ..(((.((.......)).((((((.(.((.(((....)---)).)).-)))))))......)))... ( -13.90, z-score = -2.07, R) >droWil1.scaffold_180708 2657312 65 + 12563649 AGAUGGCUAAAAAU-GAAUGGCAUUGAAGGCAGGCAAUGAAUGAAUG-AUGGCCAAAGAAUUGUAAA ...((((((...((-..((..(((((........))))).))..)).-.))))))............ ( -12.10, z-score = -1.48, R) >droGri2.scaffold_15252 14717034 63 - 17193109 -UAAAGCGACUGACUGACUUACUGAAUGAAUGAAUGAU---AGAGUAUACAGCAGUAAUAAUGCACA -....(((((((.(((...((((..((.(.....).))---..))))..))))))).....)))... ( -9.30, z-score = -1.38, R) >consensus GAGCGGCGGAGAAUAGGACAGCAGAUGGAGCAGGAUAU___UGAUUG_ACUGCUGAAACGACGUAAA ..................((((((......((.........))......))))))............ ( -5.76 = -5.23 + -0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:38 2011