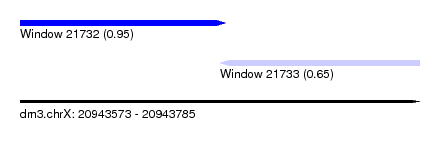
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,943,573 – 20,943,785 |
| Length | 212 |
| Max. P | 0.950332 |

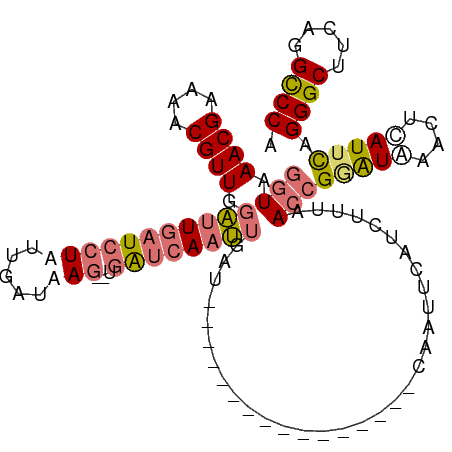
| Location | 20,943,573 – 20,943,682 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.67 |
| Shannon entropy | 0.41508 |
| G+C content | 0.38473 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -13.11 |
| Energy contribution | -14.24 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20943573 109 + 22422827 GGUAAACGGAAUCGUUGGAUUAUUCCUAUUGAUAAG--UGAUCAAUUCAUUCAUUUAACUAACUUUCAAUUCAUCCUUAACCGUGUAAACUCAUUCAGGGCUUCAGGCCCA (((.((.(((.(((..(((....)))...)))((((--(((.........)))))))................))))).)))...............((((.....)))). ( -22.30, z-score = -1.52, R) >droSim1.chrX 16101098 93 + 17042790 GGUAAACGAAAACGUUGGAUUGAUCCUUUUGUUAAG--UGAUCAAUUGAU----------------CAAUUCAUCUUUUACCGGAUAAACUUAUUCAGGGCUUCAGGCCCA ((((((.((....(((((((((((((((.....)))--.))))))))...----------------))))...)).))))))(((((....))))).((((.....)))). ( -27.20, z-score = -3.15, R) >droSec1.super_8 3231530 93 + 3762037 GGUAAACGAAAACGUUGGAUUGAUCCUUUUGUUAAG--UGAUCAAUUGAU----------------CAAUUCAUCUUUUACCGAAUAAACUCAUUCAGGGCUUCAGGUCCA ((((((.((....(((((((((((((((.....)))--.))))))))...----------------))))...)).))))))((((......)))).((((.....)))). ( -23.90, z-score = -2.32, R) >droEre2.scaffold_4690 17515135 100 + 18748788 ACUAAACGGAAACGUUGCUCUGAUCCUAUCGAUAAAGUUCGUCAAGUGAUCAAGU-----------UAACAGACUUGCAACCGGAUGAAUUCAUUUAGGGCUUCAGGCCCA ....((((....))))((.(((((((((.......((((((((..((...(((((-----------(....))))))..))..))))))))....)))))..))))))... ( -29.80, z-score = -2.77, R) >consensus GGUAAACGAAAACGUUGGAUUGAUCCUAUUGAUAAG__UGAUCAAUUGAU________________CAAUUCAUCUUUAACCGGAUAAACUCAUUCAGGGCUUCAGGCCCA (((.((((....)))).((((((((..............))))))))................................)))(((((....))))).((((.....)))). (-13.11 = -14.24 + 1.13)

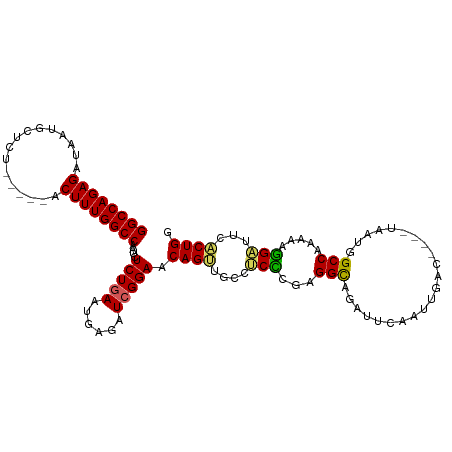
| Location | 20,943,679 – 20,943,785 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Shannon entropy | 0.32399 |
| G+C content | 0.48535 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -21.34 |
| Energy contribution | -20.90 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20943679 106 - 22422827 GGCCAGAGAUAAUGCUCU-----ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGCUUCCCGAGGCAGAUUCAAUUGACUUCUUAAUGGCCAAAAAGGAUUCACUGG ..((((.((......(((-----..(((((((((((((.....)))))..(((...((((((....)))))).)))..............))))))))..))).)).)))) ( -28.90, z-score = -0.88, R) >droSim1.chrX 16101188 102 - 17042790 GGCCAGAGAUAAUGCUCU-----ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGCCUCCCGAGGCAGAUUCAAUUGAC----UAAUGGCCAAAAAGGAUUCACUGG ..((((.((......(((-----..(((((((((((((.....)))))..(((...((((((....)))))).))).......----...))))))))..))).)).)))) ( -31.60, z-score = -1.98, R) >droSec1.super_8 3231620 102 - 3762037 GGCCAGAGAUAAUGCUCU-----ACUUUGGCCAAUCUCUGAAUGAGAUGGGAACAGUUGCCUCCCGAGGUAGAUUCAAUUGAC----UAAUGGCCAAAAAGGAUUCACUGG ..((((.((......(((-----..(((((((((((((.....)))))..(((...((((((....)))))).))).......----...))))))))..))).)).)))) ( -30.20, z-score = -1.48, R) >droYak2.chrX 20151469 106 - 21770863 GGCCAGAGAUAAUGCUCUGCUCCACUUUGGCCAAUCUCUGAAUGCGAUCGGAUCAGCUUCUUCUCGAGGCAGAUCCAACUGAC----UAAUGGCCAAAAAGGUUUC-CUGG (((.((((......)))))))((..(((((((((((.(.....).))).(((((.(((((.....))))).))))).......----...))))))))..))....-.... ( -34.20, z-score = -1.83, R) >droEre2.scaffold_4690 17515232 94 - 18748788 GGCCAGAGAUAGUGCUCUACUCUACUUUGGCCAAUCUCUGAGUGCGAUCGGAUCAGGUUCGCCUCUAGGCAG-----------------AUGGCCAACAGAGGUUUCCUGG ((((((((.(((.(.....).)))))))))))....(((((......))))).((((...((((((.(((..-----------------...)))...))))))..)))). ( -37.70, z-score = -2.32, R) >consensus GGCCAGAGAUAAUGCUCU_____ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGCCUCCCGAGGCAGAUUCAAUUGAC____UAAUGGCCAAAAAGGAUUCACUGG ((((((((................))))))))....(((((......))))).((((....(((...(((......................))).....)))...)))). (-21.34 = -20.90 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:34 2011