| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,940,119 – 20,940,212 |
| Length | 93 |
| Max. P | 0.559106 |

| Location | 20,940,119 – 20,940,212 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.02 |
| Shannon entropy | 0.59264 |
| G+C content | 0.44380 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -12.05 |
| Energy contribution | -12.87 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
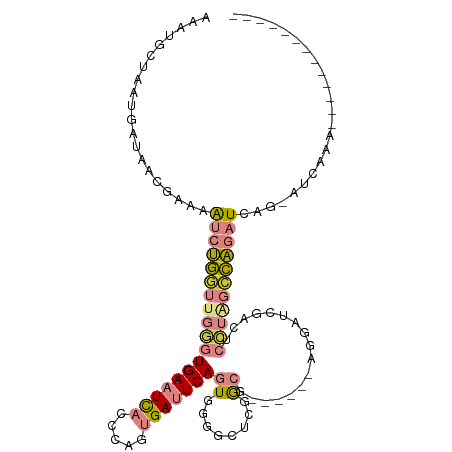
>dm3.chrX 20940119 93 + 22422827 AAAUGCUAAUGAUAACGAAAAUCUGGUUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUUGACG------AGGAUCGCCUCCUAGCCAGAUCAG-UUAAAA------------- ............((((....(((((((((((((((((((...))))))))...((((......------......)))))))))))))))..)-)))...------------- ( -30.50, z-score = -2.63, R) >droEre2.scaffold_4690 17511629 99 + 18748788 AAACAGUAAUGAUAAGGAAAGUCUGGGUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUUGGCUCCACACAGGAUCCACCCCUGGCCAGAUCAG-AUUAAA------------- .........((((..((..((...(((((((((((((((...))))))).(((((((...))))))).....))))))))))..))..)))).-......------------- ( -38.70, z-score = -2.57, R) >droYak2.chrX 20147911 93 + 21770863 AAAUGCUAAUGAUAAGGAAAGUCUGCGUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUCGGCG------AAGAUCCGCGCCUGGUCAGAUCAG-ACCAAA------------- ........................(((((((((((((((...))))))).((.(....).)).------...)))))))).(((((......)-))))..------------- ( -27.30, z-score = -0.41, R) >droSec1.super_8 3227953 93 + 3762037 AAAUGCUAAUGAUAACGAAUGUCUGGUUGGGUGAAUCACCCGGUGAUUCAGUGGGGCUCGGCG------AGGAUCGCCUCCUAGCCAGAUCUG-AUUAAA------------- ....................(((((((((((((((((((...)))))))).........((((------.....)))).)))))))))))...-......------------- ( -31.80, z-score = -1.88, R) >droSim1.chrX 16097693 93 + 17042790 AAAUGCUAAUGAUAACGAAUAUCUGGUUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUCGGCG------AGGAUCGCCUCCUAGCCAGAUCAG-AUUAAA------------- ....................(((((((((((((((((((...)))))))).........((((------.....)))).)))))))))))...-......------------- ( -31.50, z-score = -2.20, R) >dp4.chrXL_group3a 1444489 105 - 2690836 AGAAGCAAAUGAGUAAGCUAAUAAUUUCAAAUGAAUU--CAAAUGAAUCACUUCAACCCUGCC------AGAAUCGACUGCCAGAAUCUUUUGCAUCAAAGUCAGAGAUCAAA .((((....((((............))))..(((.((--(....))))))))))....(((.(------((......))).))).((((((.((......)).)))))).... ( -14.70, z-score = 0.45, R) >droPer1.super_15 1642796 105 + 2181545 AGAAGCUAAUGAGUAAGCAAAUAAUUUCAAAUGAAUU--CAAAUGAAUCACUUCAACCUUGCC------AAAAUCGACUGUCAGAAUCUUUUGCAUCAAAGUCAGAGAUCAAA .........((.(((((........((((..((....--))..))))..........))))))------).............((.(((((.((......)).)))))))... ( -10.67, z-score = 1.83, R) >consensus AAAUGCUAAUGAUAACGAAAAUCUGGUUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUCGGCG______AGGAUCGACUCCUAGCCAGAUCAG_AUCAAA_____________ ....................((((((((((((((((((.....)))))))((........)).................)))))))))))....................... (-12.05 = -12.87 + 0.82)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:31 2011