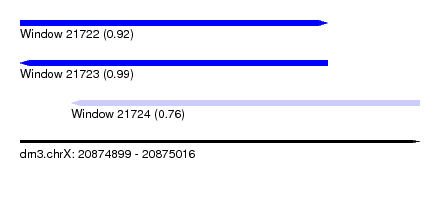
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,874,899 – 20,875,016 |
| Length | 117 |
| Max. P | 0.994748 |

| Location | 20,874,899 – 20,874,989 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.84 |
| Shannon entropy | 0.51934 |
| G+C content | 0.41255 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -11.80 |
| Energy contribution | -11.59 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20874899 90 + 22422827 -----------UAGCGACUGAGUCGACGGCCAUCGCUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUAUGGCCGCUU -----------...((((...)))).(((((((((....))(((((..(((((....))).((((((((....)))))))-----).))..))))))))))))... ( -28.30, z-score = -2.45, R) >droSim1.chrX 16034867 90 + 17042790 -----------CAGCGACUGAGACGACGGCCAUCGCUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUAUGGCCGCUU -----------.............(.(((((((((....))(((((..(((((....))).((((((((....)))))))-----).))..))))))))))))).. ( -25.90, z-score = -1.92, R) >droSec1.super_8 3157798 90 + 3762037 -----------CAGCGACUGAGACGACGGCCAUCGCUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUAUGGCCGCUU -----------.............(.(((((((((....))(((((..(((((....))).((((((((....)))))))-----).))..))))))))))))).. ( -25.90, z-score = -1.92, R) >droYak2.chrX 1683929 90 - 21770863 -----------CAGCGACUGAGACGACGGCCAUCGCUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUAUGGCCGCUU -----------.............(.(((((((((....))(((((..(((((....))).((((((((....)))))))-----).))..))))))))))))).. ( -25.90, z-score = -1.92, R) >droEre2.scaffold_4690 1300588 90 - 18748788 -----------CAGCGACUGAGACGACGGCCAUCGCUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUAUGGCCGCUU -----------.............(.(((((((((....))(((((..(((((....))).((((((((....)))))))-----).))..))))))))))))).. ( -25.90, z-score = -1.92, R) >droAna3.scaffold_13334 218036 99 - 1562580 --CAGAAAGAGACGAGACUGAGACGUGAAGCAGCGCUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUAUGGCCGCUU --.................(((.(((......))))))((((((((..(((((....))).((((((((....)))))))-----).))..))).....))))).. ( -21.60, z-score = -0.37, R) >dp4.chrXL_group1e 3036882 85 - 12523060 ---CCUUAGUUCCUUAGUUGAGGG------CAUCGUUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUAUG------- ---((((((........)))))).------((.((((((((.((((((.((((....))))))))))))))))))))...-----..............------- ( -22.70, z-score = -2.52, R) >droPer1.super_22 722166 85 + 1688296 ---CCUUAGUUCCUUAGUUGAGGG------CAUCGUUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUAUG------- ---((((((........)))))).------((.((((((((.((((((.((((....))))))))))))))))))))...-----..............------- ( -22.70, z-score = -2.52, R) >droWil1.scaffold_180702 2409210 102 + 4511350 ----CAGAGAACCCACACUGAGGGAACACCAGGCGCUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUUUGAUUGAAGUGAUUAUAUGGCCGCUG ----.......(((.......))).......((((((....(((((..(((((..((..(...((((((....)))))))..)))))))..)))))..)).)))). ( -25.10, z-score = -1.21, R) >droMoj3.scaffold_6473 4747877 90 + 16943266 -----AUAGUAACUGGCUCGAAGG-----CUC-GGAUUGCCGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUUGGACCACUU -----........(((..(((.((-----(..-.....))).((((..(((((....))).((((((((....)))))))-----).))..)))))))..)))... ( -22.10, z-score = -1.72, R) >droVir3.scaffold_12970 5186078 81 + 11907090 --------------GGCUCGAAGA-----UUU-GGAUUGGCGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUUGGACCACUU --------------((..(((...-----...-......(((((((((.((((....))))))))))))).....(((((-----.....))))))))..)).... ( -18.80, z-score = -1.75, R) >droGri2.scaffold_15081 4104858 96 - 4274704 CAACUACAAUAAGGAGCCCGAAGG-----UUUCGGAUUCACGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU-----GAAGUGAUUAUUGGGGCACUU ....((((((..((((((....))-----))))..)))...)))..(((((((((((((..((((((((....)))))))-----)...)))))).)))))))... ( -23.10, z-score = -1.83, R) >consensus ___________CAGCGACUGAGGCGAC__CCAUCGCUUGCGGUAAUUGCUUCAUAAUUGAACAAUUAUGCAAACGUGAUU_____GAAGUGAUUAUAUGGCCGCUU .................................((....))(((((..((((((((((....))))))((....)).........))))..))))).......... (-11.80 = -11.59 + -0.21)

| Location | 20,874,899 – 20,874,989 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 74.84 |
| Shannon entropy | 0.51934 |
| G+C content | 0.41255 |
| Mean single sequence MFE | -21.84 |
| Consensus MFE | -11.16 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20874899 90 - 22422827 AAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCGUCGACUCAGUCGCUA----------- ..((((((((...........-----.....(((((((.((((((((((....)).))))))))..)))))))))))))))((((...))))...----------- ( -28.09, z-score = -3.70, R) >droSim1.chrX 16034867 90 - 17042790 AAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCGUCGUCUCAGUCGCUG----------- .(((((((((...........-----.....(((((((.((((((((((....)).))))))))..)))))))))))))(.(......).)))).----------- ( -26.39, z-score = -3.17, R) >droSec1.super_8 3157798 90 - 3762037 AAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCGUCGUCUCAGUCGCUG----------- .(((((((((...........-----.....(((((((.((((((((((....)).))))))))..)))))))))))))(.(......).)))).----------- ( -26.39, z-score = -3.17, R) >droYak2.chrX 1683929 90 + 21770863 AAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCGUCGUCUCAGUCGCUG----------- .(((((((((...........-----.....(((((((.((((((((((....)).))))))))..)))))))))))))(.(......).)))).----------- ( -26.39, z-score = -3.17, R) >droEre2.scaffold_4690 1300588 90 + 18748788 AAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCGUCGUCUCAGUCGCUG----------- .(((((((((...........-----.....(((((((.((((((((((....)).))))))))..)))))))))))))(.(......).)))).----------- ( -26.39, z-score = -3.17, R) >droAna3.scaffold_13334 218036 99 + 1562580 AAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGCUGCUUCACGUCUCAGUCUCGUCUCUUUCUG-- (((((((..............-----......((((((.((((((((((....)).))))))))..))))))))))))).........................-- ( -19.05, z-score = -1.70, R) >dp4.chrXL_group1e 3036882 85 + 12523060 -------CAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAACGAUG------CCCUCAACUAAGGAACUAAGG--- -------..............-----.....(((((((.((((((((((....)).))))))))..)))))))...------.(((......)))........--- ( -17.40, z-score = -3.08, R) >droPer1.super_22 722166 85 - 1688296 -------CAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAACGAUG------CCCUCAACUAAGGAACUAAGG--- -------..............-----.....(((((((.((((((((((....)).))))))))..)))))))...------.(((......)))........--- ( -17.40, z-score = -3.08, R) >droWil1.scaffold_180702 2409210 102 - 4511350 CAGCGGCCAUAUAAUCACUUCAAUCAAAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGCCUGGUGUUCCCUCAGUGUGGGUUCUCUG---- (((.((((.......((((........(((((((((((.((((((((((....)).))))))))..)))))))..))))........))))..)).)).)))---- ( -23.59, z-score = -1.26, R) >droMoj3.scaffold_6473 4747877 90 - 16943266 AAGUGGUCCAAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACGGCAAUCC-GAG-----CCUUCGAGCCAGUUACUAU----- ((.(((..(............-----......((((.(((((((....)))))))))))......(((.....-..)-----))...)..))).)).....----- ( -17.70, z-score = -1.52, R) >droVir3.scaffold_12970 5186078 81 - 11907090 AAGUGGUCCAAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACGCCAAUCC-AAA-----UCUUCGAGCC-------------- (((((((......))))))).-----......(((.((.((((((((((....)).)))))))).)).)))..-...-----..........-------------- ( -12.80, z-score = -1.51, R) >droGri2.scaffold_15081 4104858 96 + 4274704 AAGUGCCCCAAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACGUGAAUCCGAAA-----CCUUCGGGCUCCUUAUUGUAGUUG ....((..((((((.......-----..((((((((((((((((....))))))...))))..)))))).(((((..-----...)))))....))))))..)).. ( -20.50, z-score = -2.28, R) >consensus AAGCGGCCAUAUAAUCACUUC_____AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGG__GUCGCCUCAGUCGCGG___________ ................................((((((.((((((((((....)).))))))))..)))))).................................. (-11.16 = -11.57 + 0.40)


| Location | 20,874,914 – 20,875,016 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Shannon entropy | 0.43555 |
| G+C content | 0.41830 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -11.16 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20874914 102 - 22422827 -------------GGGAAACGCACAUAAACAGAGGCGCAUAAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCG -------------.(....)((.(((.......(((((....)).))).............-----.....(((((((.((((((((((....)).))))))))..)))))))))))).. ( -27.00, z-score = -2.25, R) >droSim1.chrX 16034882 102 - 17042790 -------------GGGAAACGCACAUAUACAGAGGCGCAUAAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCG -------------.(....)((.(((.......(((((....)).))).............-----.....(((((((.((((((((((....)).))))))))..)))))))))))).. ( -27.00, z-score = -2.17, R) >droSec1.super_8 3157813 102 - 3762037 -------------GGGAAACGCACAUAUACAGAGGCGCAUAAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCG -------------.(....)((.(((.......(((((....)).))).............-----.....(((((((.((((((((((....)).))))))))..)))))))))))).. ( -27.00, z-score = -2.17, R) >droYak2.chrX 1683944 102 + 21770863 -------------GGAAAACACACAUAAACAGAGGCGCAUAAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCG -------------.....................((......))((((((...........-----.....(((((((.((((((((((....)).))))))))..))))))))))))). ( -25.39, z-score = -2.64, R) >droEre2.scaffold_4690 1300603 102 + 18748788 -------------GGAAAACACACAUAAACAGAGGCGCAUAAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCG -------------.....................((......))((((((...........-----.....(((((((.((((((((((....)).))))))))..))))))))))))). ( -25.39, z-score = -2.64, R) >droAna3.scaffold_13334 218060 115 + 1562580 GGGAACUGGGAACUCGGAAAGCACAUAAACAGAGGCGCAUAAGCGGCCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGCUGCUUC .....((((....)))).(((((..........(((((....)).))).............-----.....(((((((.((((((((((....)).))))))))..))))))).))))). ( -27.60, z-score = -1.13, R) >dp4.chrXL_group1e 3036900 86 + 12523060 --------------------------ACACAGAGAAACA--GGGGGGCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAACGAUGCCC- --------------------------.............--...((((((...........-----.....(((((((.((((((((((....)).))))))))..)))))))))))))- ( -21.49, z-score = -2.92, R) >droPer1.super_22 722184 84 - 1688296 --------------------------ACACACAGAAACA--G--GGGCAUAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAACGAUGCCC- --------------------------.............--.--((((((...........-----.....(((((((.((((((((((....)).))))))))..)))))))))))))- ( -21.49, z-score = -3.91, R) >droWil1.scaffold_180702 2409232 106 - 4511350 --------------GGGAAAAACAGUAAACGGAGAAACAUCAGCGGCCAUAUAAUCACUUCAAUCAAAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGCCUGGUG --------------.......................((((((.((........))...............(((((((.((((((((((....)).))))))))..))))))).)))))) ( -18.90, z-score = -0.00, R) >droMoj3.scaffold_6473 4747893 101 - 16943266 -------------GUGACGAGGACAUAAACAUUGGCGCCUAAGUGGUCCAAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACGGCAAUCCGAGCCU- -------------((((.((((........(((((.(((.....)))))))).....))))-----..))))((((.(((((((....)))))))))))......(((.......))).- ( -24.12, z-score = -1.43, R) >droVir3.scaffold_12970 5186085 101 - 11907090 -------------GUGACGAGGGCAUAAACAUUGUCGCCUAAGUGGUCCAAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACGCCAAUCCAAAUCU- -------------((((...(((((((.....))).))))(((((((......))))))).-----..))))(((.((.((((((((((....)).)))))))).)).)))........- ( -21.80, z-score = -1.81, R) >droGri2.scaffold_15081 4104879 102 + 4274704 -------------GUGAAGAGGACAUAAACACUGCCGCCUAAGUGCCCCAAUAAUCACUUC-----AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACGUGAAUCCGAAACCU -------------.(((((.((.......((((........)))).........)).))))-----).((((((((((((((((....))))))...))))..))))))........... ( -18.29, z-score = -1.10, R) >consensus _____________GGGAAAAGCACAUAAACAGAGGCGCAUAAGCGGCCAUAUAAUCACUUC_____AAUCACGUUUGCAUAAUUGUUCAAUUAUGAAGCAAUUACCGCAAGCGAUGGCCG ........................................................................((((((.((((((((((....)).))))))))..))))))........ (-11.16 = -11.57 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:27 2011