| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,872,105 – 20,872,182 |
| Length | 77 |
| Max. P | 0.697990 |

| Location | 20,872,105 – 20,872,182 |
|---|---|
| Length | 77 |
| Sequences | 11 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 95.16 |
| Shannon entropy | 0.09753 |
| G+C content | 0.33318 |
| Mean single sequence MFE | -16.49 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.20 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
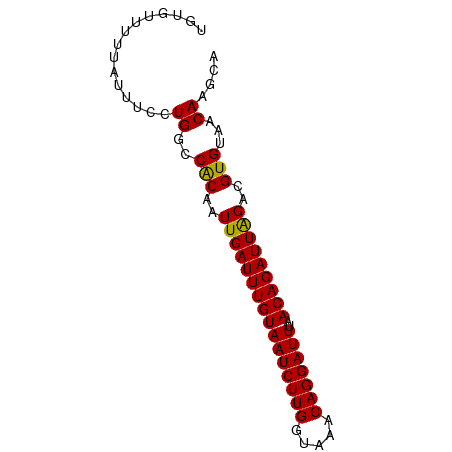
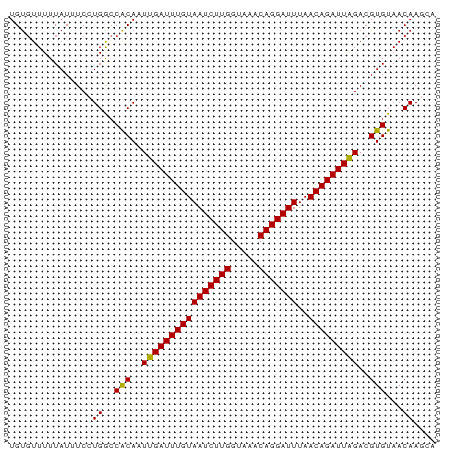
>dm3.chrX 20872105 77 + 22422827 UGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCA ...............((..(((..((((((((((((((((.....)))))))..)))))))))..)))...)).... ( -15.60, z-score = -0.89, R) >droSec1.super_8 3154996 77 + 3762037 UGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCA ...............((..(((..((((((((((((((((.....)))))))..)))))))))..)))...)).... ( -15.60, z-score = -0.89, R) >droYak2.chrX 20084768 77 + 21770863 UGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCA ...............((..(((..((((((((((((((((.....)))))))..)))))))))..)))...)).... ( -15.60, z-score = -0.89, R) >droEre2.scaffold_4690 17446514 77 + 18748788 UGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCA ...............((..(((..((((((((((((((((.....)))))))..)))))))))..)))...)).... ( -15.60, z-score = -0.89, R) >droAna3.scaffold_13334 214881 77 - 1562580 UGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCA ...............((..(((..((((((((((((((((.....)))))))..)))))))))..)))...)).... ( -15.60, z-score = -0.89, R) >droPer1.super_22 720722 77 + 1688296 UGUGUUUUUAUUUCCUGGGCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCA (((((((.........))))))).((((((((((((((((.....)))))))..))))))))).............. ( -17.60, z-score = -1.63, R) >dp4.chrXL_group1e 3035554 77 - 12523060 UGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCA ...............((..(((..((((((((((((((((.....)))))))..)))))))))..)))...)).... ( -15.60, z-score = -0.89, R) >droWil1.scaffold_180702 2410496 73 + 4511350 UGUGUUUUUAUUUC-UGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAA--- ..............-((..(((..((((((((((((((((.....)))))))..)))))))))..)))...)).--- ( -15.60, z-score = -1.45, R) >droMoj3.scaffold_6473 4745185 73 + 16943266 UGUGUUUUUAUGUC-UGACCGCAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAA--- ..((((..((((((-(((.(((((.....))))(((((((.....))))))).....).))))))))).)))).--- ( -19.30, z-score = -2.69, R) >droVir3.scaffold_12970 5184210 73 + 11907090 UGUGUUUUUAUGUC-UGACCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAA--- ..((((..((((((-(((.(((((.....))))(((((((.....))))))).....).))))))))).)))).--- ( -19.00, z-score = -2.44, R) >droGri2.scaffold_15081 4102867 73 - 4274704 UGCUUUUUUAUGUC-UGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUGGACGUGUAACAA--- ........((((((-((....)....((((((((((((((.....)))))))..))))))))))))))......--- ( -16.30, z-score = -1.48, R) >consensus UGUGUUUUUAUUUCCUGGCCACAAUUGAUUUGUAAUCUUGGUAAACAGGAUUUAACAGAUUAGACGUGUAACAAGCA ...............((..(((..((((((((((((((((.....)))))))..)))))))))..)))...)).... (-15.36 = -15.20 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:24 2011