| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,847,273 – 20,847,378 |
| Length | 105 |
| Max. P | 0.984697 |

| Location | 20,847,273 – 20,847,378 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Shannon entropy | 0.37089 |
| G+C content | 0.54917 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -27.21 |
| Energy contribution | -28.29 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
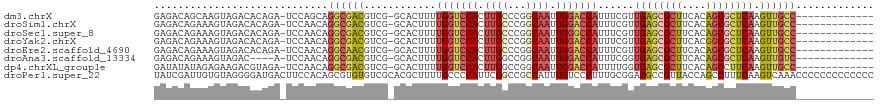
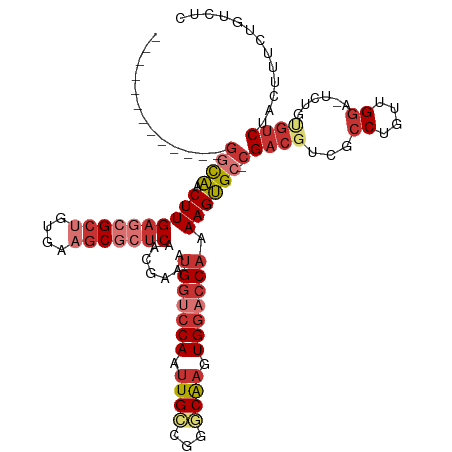
>dm3.chrX 20847273 105 + 22422827 GAGACAGCAAGUAGACACAGA-UCCAGCAGGCGACGUCG-GCACUUUUGGUCCACUUGCCCGGCAAUUGGACCAUUUCGUUGAGCGCUUCACAGCGCUCAAGUUGCC------------- ..(((.((..((.((......-))..))..))...)))(-(((....(((((((.((((...)))).))))))).....(((((((((....)))))))))..))))------------- ( -37.90, z-score = -2.57, R) >droSim1.chrX 16015787 105 + 17042790 GAGACAGAAAGUAGACACAGA-UCCAACAGGCGACGUCG-GCACUUUUGGUCCACUUGCCCGGCAAUUGGACCAUUUCGUUGAGCGCUUCACAGCGCUCAAGUUGCC------------- .....................-.......((((((....-.......(((((((.((((...)))).))))))).....(((((((((....)))))))))))))))------------- ( -37.60, z-score = -2.97, R) >droSec1.super_8 3130350 105 + 3762037 GAGACAGAAAGUAGACACAGA-UCCAACAGGCGACGUCG-GCACUUUUGGUCCACUUGCCCGGCAAUUGGGCCAUUUCGUUGAGCGCUUCACAGCGCUCAAGUUGCC------------- .....................-.......((((((....-.......(((((((.((((...)))).))))))).....(((((((((....)))))))))))))))------------- ( -37.00, z-score = -2.29, R) >droYak2.chrX 20058716 105 + 21770863 GAGACAGAAAGUAGACACAGA-UCCAACAGGCGACGUCG-GCACUUUUGGUCCACUUGCCCGGCAAUUGGACCAUUUCGUUGAGCGCUUCACGGCGCUCAAGUUGCC------------- .....................-.......((((((....-.......(((((((.((((...)))).))))))).....(((((((((....)))))))))))))))------------- ( -37.60, z-score = -2.51, R) >droEre2.scaffold_4690 17420945 105 + 18748788 GAGACAGAAAGUAGACACAGA-UCCAACAGGCAACGUCG-GCACUUUUGGUCCACUUGCCCGGCAAUUGGACCAUUUCGUUGAGCGCUUCACAGCGCUCAAGUUGCC------------- .....................-.......((((((....-.......(((((((.((((...)))).))))))).....(((((((((....)))))))))))))))------------- ( -37.90, z-score = -3.39, R) >droAna3.scaffold_13334 1381587 101 + 1562580 GAGACAGAAAGUAGAC----A-UCCAACAGGCGACGUCG-GCACUUUUGGUCCACUUGGCCGGCAAUUGGACCAUUUCGGUGAGCGCUUCACAGCGCUCAAGUUGUC------------- ..((((((((((.(((----.-((........)).))).-)).....(((((((.(((.....))).)))))))))))..((((((((....))))))))...))))------------- ( -35.10, z-score = -1.98, R) >dp4.chrXL_group1e 9482001 105 + 12523060 GAUAUAUAGAGAAGACGUAGA-UCCAACAGGCGACGUCG-GCACUUUUGGUCCACUUGGCCGGCAAUUGGACCAUUUUGGUGAGCGCUUCACAGCCUUCAAGUUGCC------------- .............(((((...-.((....))..)))))(-((((((.(((((((.(((.....))).))))))).......(((.(((....))).)))))).))))------------- ( -29.90, z-score = -0.68, R) >droPer1.super_22 962615 120 - 1688296 UAUCGAUUGUGUAGGGGAUGACUUCCACAGCGUGUGUCGCACGCUUUUGCCCCAUUCGGCCGCCAUUUGUCCCUUUGCGGAGGCCGUUACCAGCCUUUCAAGUCAAACCCCCCCCCCCCC .............((((.((((((....((((((.....))))))...((......(((((.((..............)).)))))......)).....))))))..))))......... ( -34.04, z-score = -1.26, R) >consensus GAGACAGAAAGUAGACACAGA_UCCAACAGGCGACGUCG_GCACUUUUGGUCCACUUGCCCGGCAAUUGGACCAUUUCGUUGAGCGCUUCACAGCGCUCAAGUUGCC_____________ .............................((((((............(((((((.((((...)))).)))))))......((((((((....)))))))).))))))............. (-27.21 = -28.29 + 1.08)

| Location | 20,847,273 – 20,847,378 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.95 |
| Shannon entropy | 0.37089 |
| G+C content | 0.54917 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -23.91 |
| Energy contribution | -26.09 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.984055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
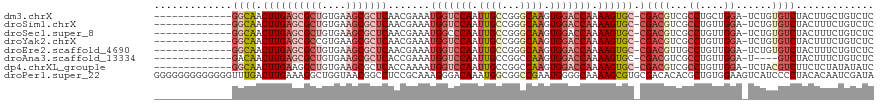
>dm3.chrX 20847273 105 - 22422827 -------------GGCAACUUGAGCGCUGUGAAGCGCUCAACGAAAUGGUCCAAUUGCCGGGCAAGUGGACCAAAAGUGC-CGACGUCGCCUGCUGGA-UCUGUGUCUACUUGCUGUCUC -------------((((.((((((((((....))))))).......(((((((.((((...)))).))))))).))))))-)((((..((..(.((((-(....))))))..)))))).. ( -42.90, z-score = -3.29, R) >droSim1.chrX 16015787 105 - 17042790 -------------GGCAACUUGAGCGCUGUGAAGCGCUCAACGAAAUGGUCCAAUUGCCGGGCAAGUGGACCAAAAGUGC-CGACGUCGCCUGUUGGA-UCUGUGUCUACUUUCUGUCUC -------------((((.((((((((((....))))))).......(((((((.((((...)))).))))))).))))))-)((((...((....)).-....))))............. ( -40.40, z-score = -2.93, R) >droSec1.super_8 3130350 105 - 3762037 -------------GGCAACUUGAGCGCUGUGAAGCGCUCAACGAAAUGGCCCAAUUGCCGGGCAAGUGGACCAAAAGUGC-CGACGUCGCCUGUUGGA-UCUGUGUCUACUUUCUGUCUC -------------((((..(((((((((....))))))))).((((..((((.......))))..((((((....((..(-(((((.....)))))).-.))..)))))))))))))).. ( -40.60, z-score = -2.83, R) >droYak2.chrX 20058716 105 - 21770863 -------------GGCAACUUGAGCGCCGUGAAGCGCUCAACGAAAUGGUCCAAUUGCCGGGCAAGUGGACCAAAAGUGC-CGACGUCGCCUGUUGGA-UCUGUGUCUACUUUCUGUCUC -------------((((.(((((((((......)))))).......(((((((.((((...)))).))))))).))))))-)((((...((....)).-....))))............. ( -38.10, z-score = -2.05, R) >droEre2.scaffold_4690 17420945 105 - 18748788 -------------GGCAACUUGAGCGCUGUGAAGCGCUCAACGAAAUGGUCCAAUUGCCGGGCAAGUGGACCAAAAGUGC-CGACGUUGCCUGUUGGA-UCUGUGUCUACUUUCUGUCUC -------------(((((((((((((((....))))))))).....(((((((.((((...)))).))))))).......-....))))))...((((-(....)))))........... ( -41.20, z-score = -3.31, R) >droAna3.scaffold_13334 1381587 101 - 1562580 -------------GACAACUUGAGCGCUGUGAAGCGCUCACCGAAAUGGUCCAAUUGCCGGCCAAGUGGACCAAAAGUGC-CGACGUCGCCUGUUGGA-U----GUCUACUUUCUGUCUC -------------((((...((((((((....))))))))......(((((((.(((.....))).)))))))((((((.-.((((((........))-)----))))))))).)))).. ( -39.10, z-score = -3.64, R) >dp4.chrXL_group1e 9482001 105 - 12523060 -------------GGCAACUUGAAGGCUGUGAAGCGCUCACCAAAAUGGUCCAAUUGCCGGCCAAGUGGACCAAAAGUGC-CGACGUCGCCUGUUGGA-UCUACGUCUUCUCUAUAUAUC -------------((((.(((((..(((....)))..)).......(((((((.(((.....))).))))))).))))))-)(((((..((....)).-...)))))............. ( -31.80, z-score = -1.56, R) >droPer1.super_22 962615 120 + 1688296 GGGGGGGGGGGGGUUUGACUUGAAAGGCUGGUAACGGCCUCCGCAAAGGGACAAAUGGCGGCCGAAUGGGGCAAAAGCGUGCGACACACGCUGUGGAAGUCAUCCCCUACACAAUCGAUA ((..(...(((((..((((((...((((((....))))))((((................(((......)))...((((((.....)))))))))))))))).)))))...)..)).... ( -45.90, z-score = -1.72, R) >consensus _____________GGCAACUUGAGCGCUGUGAAGCGCUCAACGAAAUGGUCCAAUUGCCGGGCAAGUGGACCAAAAGUGC_CGACGUCGCCUGUUGGA_UCUGUGUCUACUUUCUGUCUC .............(((((((((((((((....))))))).......(((((((.((((...)))).))))))).))))....((((...((....))......)))).......)))).. (-23.91 = -26.09 + 2.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:20 2011