| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,843,985 – 20,844,090 |
| Length | 105 |
| Max. P | 0.531999 |

| Location | 20,843,985 – 20,844,090 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.82 |
| Shannon entropy | 0.34166 |
| G+C content | 0.40408 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.531999 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 20843985 105 - 22422827 GUCGAGCCUCACUCUUUGUUUUCUCGUUUUUGUGUUCAGCAAUAGCAUC-AAAAUUGCAAAGAAAAGCCAGCAAACAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ....(((.....(((((((......(((((.(((((.(....)))))).-))))).)))))))...((((((..(((((.........))))).)))))).))).. ( -24.80, z-score = -0.91, R) >droSim1.chrX 16012513 105 - 17042790 GUCGGGCCUCACUCUUUGUUUUCUCGUUUUUGUGUUCAGCAAUAGCAUC-AAAAUUGCAAAGAAAAGCCAGCAAACAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ....(((.....(((((((......(((((.(((((.(....)))))).-))))).)))))))...((((((..(((((.........))))).)))))).))).. ( -24.80, z-score = -0.70, R) >droSec1.super_8 3127032 105 - 3762037 GUCGGGCCUCACUCUUUGUUUUCUCGUUUUUGUGUUCAGCAAUAGCAUC-AAAAUUGCAAAGAAAAGCCAGCAAACAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ....(((.....(((((((......(((((.(((((.(....)))))).-))))).)))))))...((((((..(((((.........))))).)))))).))).. ( -24.80, z-score = -0.70, R) >droYak2.chrX 20055408 105 - 21770863 GUCGGGCCGCACUUUUGGUUUUCUCCUUUUUGUGUUCAGCAAUAGCAUC-AAAAUUGCAAAGAAAAGCCAGCAAACAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ........((((....((......)).....)))).((((....(((..-.....)))........((((((..(((((.........))))).)))))).)))). ( -25.50, z-score = -0.24, R) >droEre2.scaffold_4690 17417700 105 - 18748788 GUCGGGCCUCACUUUUUGUUUUCUCCAUUUUGUGUUCAGCAAUAGCAUC-AAAAUUGCAAAGAAAAGCCAGCAAACAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ...(((....((.....))....)))..........((((....(((..-.....)))........((((((..(((((.........))))).)))))).)))). ( -22.60, z-score = -0.05, R) >droAna3.scaffold_13334 177234 88 + 1562580 ------------------ACUGUUUCUUUUUGUGUUGAGCAAUAGCUUCAAAAAUGGCAAAGAAAAGCCGGCAAACAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ------------------((((...((.(((((.((((((....)).))))...((((........))))))))).)).))))........(((((.....))))) ( -21.30, z-score = -0.23, R) >droPer1.super_11308 363 105 + 930 UCCAGUUCUCUCUCUCUCGUUUUUUUUUUUCGUGUUCUGCAAUAGCAUC-AAAAUUGCAAAGAAAAGCCAGCAAACAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ..(((((...................(((((......((((((......-...))))))..)))))((((((..(((((.........))))).))))))))))). ( -21.60, z-score = -1.08, R) >dp4.chrXL_group1e 9479305 102 - 12523060 UUCAGUUCUCUCUCUC---GUUUUUUUUUUUGUGUUCUGCAAUAGCAUC-AAAAUUGCAAAGAAAAGCCAGCAAACAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ..(((((.........---....(((((((((((...(((....)))..-.....)))))))))))((((((..(((((.........))))).))))))))))). ( -23.40, z-score = -1.41, R) >droWil1.scaffold_180702 2063562 82 + 4511350 ---------------------CUUUUUUUUCCAGU--AGCAAUAGCAUC-AAAAUUGCAAAGAAAAGCCAGCAAGCAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ---------------------............((--(((....(((..-.....)))........((((((..(((((.........))))).)))))).))))) ( -22.30, z-score = -2.61, R) >consensus GUCGGGCCUCACUCUUUGUUUUCUCCUUUUUGUGUUCAGCAAUAGCAUC_AAAAUUGCAAAGAAAAGCCAGCAAACAGUCAGUAAAAUAUUGUAGCUGGCAGCUGC ....................................((((....(((........)))........((((((..(((((.........))))).)))))).)))). (-19.45 = -19.50 + 0.05)

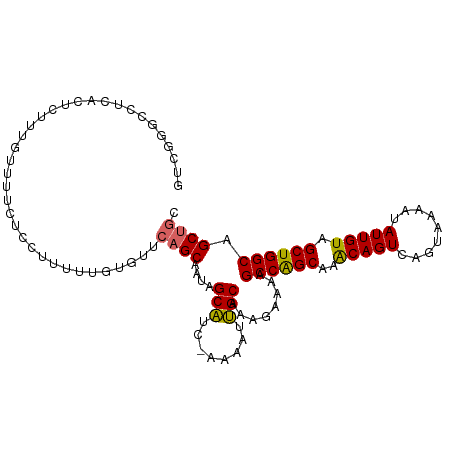

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:18 2011