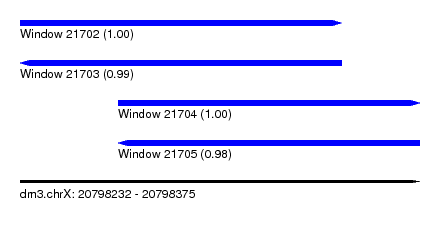
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,798,232 – 20,798,375 |
| Length | 143 |
| Max. P | 0.998569 |

| Location | 20,798,232 – 20,798,347 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
| Shannon entropy | 0.39658 |
| G+C content | 0.48169 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -29.75 |
| Energy contribution | -29.66 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.998569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
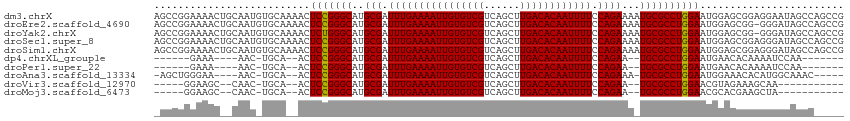
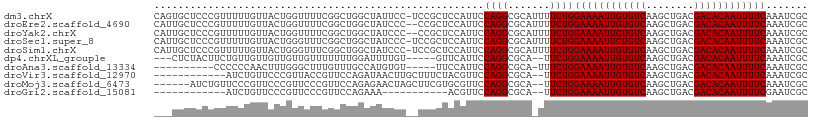
>dm3.chrX 20798232 115 + 22422827 AGCCGGAAAACUGCAAUGUGCAAAACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGGAGGAAUAGCCAGCCG ...(((....((((...((.....))(((((((..(((.((((((((((((((((......)))))))))))))...))).)))))))))).....)))).((.....))..))) ( -38.50, z-score = -1.98, R) >droEre2.scaffold_4690 17380201 114 + 18748788 AGCCGGAAAACUGCAAUGUGCAAAACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGG-GGGAUAGCCAGCCG .((.((....((((...((.....))(((((((..(((.((((((((((((((((......)))))))))))))...))).)))))))))).....))))-.......)).)).. ( -38.70, z-score = -1.58, R) >droYak2.chrX 20014847 114 + 21770863 AGCCGGAAAACUGCAAUGUGCAAAACUCUGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGG-GGGAUAGCCAGCCG .((.((....((((...((.....))((..(((..(((.((((((((((((((((......)))))))))))))...))).))))))..)).....))))-.......)).)).. ( -36.10, z-score = -1.00, R) >droSec1.super_8 3060448 115 + 3762037 AGCCGGAAAACUGCAAUGUGCAAAACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGGAGGGAUAGCCAGCCG ..(((......(((.....)))....(((((((..(((.((((((((((((((((......)))))))))))))...))).)))))))))).)))..(((..((....))..))) ( -39.60, z-score = -2.03, R) >droSim1.chrX 15966750 115 + 17042790 AGCCGGAAAACUGCAAUGUGCAAAACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGGAGGGAUAGCCAGCCG ..(((......(((.....)))....(((((((..(((.((((((((((((((((......)))))))))))))...))).)))))))))).)))..(((..((....))..))) ( -39.60, z-score = -2.03, R) >dp4.chrXL_group1e 9445988 93 + 12523060 ------GAAA----AAC-UGCA--ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAA--UGCGCCUGGAAUGAACACAAAAUCCAA------- ------....----...-....--..(((((((..(((.((((((((((((((((......)))))))))))).)))).--)))))))))).................------- ( -32.00, z-score = -4.47, R) >droPer1.super_22 927852 93 - 1688296 ------GAAA----AAC-UGCA--ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAA--UGCGCCUGGAAUGAACACAAAAUCCAA------- ------....----...-....--..(((((((..(((.((((((((((((((((......)))))))))))).)))).--)))))))))).................------- ( -32.00, z-score = -4.47, R) >droAna3.scaffold_13334 135778 101 - 1562580 -AGCUGGGAA----AAC-UGCA--ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAA-UGCGCCUGGAAUGGAAACACAUGGCAAAC----- -.(((.....----...-....--..(((((((..(((.((((((((((((((((......)))))))))))))..))).-)))))))))).((....))...)))....----- ( -36.10, z-score = -3.77, R) >droVir3.scaffold_12970 5123422 92 + 11907090 -----GGAAGC--CAAC-UGCA--ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAA--UGCGCCUGGAACGUAGAAAGCAA----------- -----....((--...(-(((.--..(((((((..(((.((((((((((((((((......)))))))))))).)))).--))))))))))..))))...))..----------- ( -37.50, z-score = -4.67, R) >droMoj3.scaffold_6473 4661571 92 + 16943266 -----GGAAGC--CAAC-UGCA--ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAA--UGCGCCUGGAACGCACGAAGCUA----------- -----...(((--....-(((.--..(((((((..(((.((((((((((((((((......)))))))))))).)))).--))))))))))..)))....))).----------- ( -37.10, z-score = -4.36, R) >consensus _GCCGGAAAAC__CAAC_UGCA__ACUCCGGGCAUGCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAA_UGCGCCUGGAAUGGAGCGAAGGGAUAGCC_____ ..........................(((((((..(((.((((((((((((((((......)))))))))))).))))...))))))))))........................ (-29.75 = -29.66 + -0.09)

| Location | 20,798,232 – 20,798,347 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.52 |
| Shannon entropy | 0.39658 |
| G+C content | 0.48169 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.14 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20798232 115 - 22422827 CGGCUGGCUAUUCCUCCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGUUUUGCACAUUGCAGUUUUCCGGCU .(((..((.........((.((......)).))........((((((((((((........)))))))))))).....))..)))(((((..((((.....))))..)))))... ( -39.30, z-score = -2.91, R) >droEre2.scaffold_4690 17380201 114 - 18748788 CGGCUGGCUAUCCC-CCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGUUUUGCACAUUGCAGUUUUCCGGCU .(((..((......-..((.((......)).))........((((((((((((........)))))))))))).....))..)))(((((..((((.....))))..)))))... ( -39.30, z-score = -2.82, R) >droYak2.chrX 20014847 114 - 21770863 CGGCUGGCUAUCCC-CCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCAGAGUUUUGCACAUUGCAGUUUUCCGGCU .(((((((......-..((.((......)).))........((((((((((((........)))))))))))).....)).......(((..((((.....))))..)))))))) ( -34.00, z-score = -1.66, R) >droSec1.super_8 3060448 115 - 3762037 CGGCUGGCUAUCCCUCCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGUUUUGCACAUUGCAGUUUUCCGGCU .(((..((.........((.((......)).))........((((((((((((........)))))))))))).....))..)))(((((..((((.....))))..)))))... ( -39.30, z-score = -2.85, R) >droSim1.chrX 15966750 115 - 17042790 CGGCUGGCUAUCCCUCCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGUUUUGCACAUUGCAGUUUUCCGGCU .(((..((.........((.((......)).))........((((((((((((........)))))))))))).....))..)))(((((..((((.....))))..)))))... ( -39.30, z-score = -2.85, R) >dp4.chrXL_group1e 9445988 93 - 12523060 -------UUGGAUUUUGUGUUCAUUCCAGGCGCA--UUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU--UGCA-GUU----UUUC------ -------..........(((..(((((.((((((--((...((((((((((((........)))))))))))).))).))..))).)))))--.)))-...----....------ ( -28.50, z-score = -2.24, R) >droPer1.super_22 927852 93 + 1688296 -------UUGGAUUUUGUGUUCAUUCCAGGCGCA--UUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU--UGCA-GUU----UUUC------ -------..........(((..(((((.((((((--((...((((((((((((........)))))))))))).))).))..))).)))))--.)))-...----....------ ( -28.50, z-score = -2.24, R) >droAna3.scaffold_13334 135778 101 + 1562580 -----GUUUGCCAUGUGUUUCCAUUCCAGGCGCA-UUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU--UGCA-GUU----UUCCCAGCU- -----..........(((....(((((.((((..-......((((((((((((........))))))))))))........)))).)))))--.)))-...----.........- ( -27.89, z-score = -1.45, R) >droVir3.scaffold_12970 5123422 92 - 11907090 -----------UUGCUUUCUACGUUCCAGGCGCA--UUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU--UGCA-GUUG--GCUUCC----- -----------..(((..((...((((.((((((--((...((((((((((((........)))))))))))).))).))..))).)))).--...)-)..)--))....----- ( -29.60, z-score = -2.28, R) >droMoj3.scaffold_6473 4661571 92 - 16943266 -----------UAGCUUCGUGCGUUCCAGGCGCA--UUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU--UGCA-GUUG--GCUUCC----- -----------.((((...(((..(((.((((((--((...((((((((((((........)))))))))))).))).))..))).)))..--.)))-...)--)))...----- ( -31.80, z-score = -2.31, R) >consensus _____GGCUAUCCCUCCGCUCCAUUCCAGGCGCA_UUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGCAUGCCCGGAGU__UGCA_AUUG__GUUUCCCGGC_ ......................(((((.((((.........((((((((((((........))))))))))))........)))).)))))........................ (-25.20 = -25.14 + -0.06)

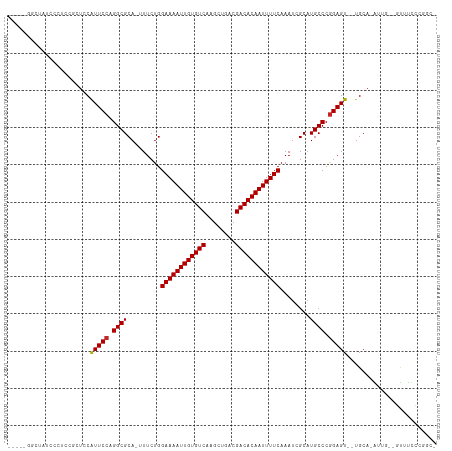
| Location | 20,798,267 – 20,798,375 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.89 |
| Shannon entropy | 0.53866 |
| G+C content | 0.46480 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -19.09 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20798267 108 + 22422827 GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGGA-GGAAUAGCCAGCCGAAAACCAGUAACAAAAACGGGAGCACUG .......(((((((((((((......)))))))))))))(((....(((.((((...(((..(((.-((.....))..)))....)))..........)))).)))))) ( -30.62, z-score = -1.81, R) >droEre2.scaffold_4690 17380236 107 + 18748788 GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGG--GGGAUAGCCAGCCGAAAACCAGUAACAAAAACGGGAGCAAUG .......(((((((((((((......))))))))))))).......(((.((((...(((..(((--.((....))..)))....)))..........)))).)))... ( -32.12, z-score = -1.96, R) >droYak2.chrX 20014882 107 + 21770863 GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGG--GGGAUAGCCAGCCGAAAACCAGUAACAAAAACGGGAGCAAUG .......(((((((((((((......))))))))))))).......(((.((((...(((..(((--.((....))..)))....)))..........)))).)))... ( -32.12, z-score = -1.96, R) >droSec1.super_8 3060483 108 + 3762037 GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGGA-GGGAUAGCCAGCCGAAACCCAGUAACAAAAACGGGAGCAAUG .......(((((((((((((......))))))))))))).......(((.((((...(((..(((.-.((....))..)))....)))..........)))).)))... ( -33.62, z-score = -2.45, R) >droSim1.chrX 15966785 108 + 17042790 GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAAAUGCGCCUGGAAUGGAGCGGA-GGGAUAGCCAGCCGAAACCCAGUAACAAAAACGGGAGCAAUG .......(((((((((((((......))))))))))))).......(((.((((...(((..(((.-.((....))..)))....)))..........)))).)))... ( -33.62, z-score = -2.45, R) >dp4.chrXL_group1e 9446010 99 + 12523060 GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAA--UGCGCCUGGAAUGAAC-----ACAAAAUCCAAAAAAACAACAACAACAACAGAAGUAGAG--- ((((((((((((((((((((......)))))))))))))..)))--).))).((((.(....-----....).)))).............................--- ( -22.70, z-score = -2.71, R) >droAna3.scaffold_13334 135805 93 - 1562580 GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAA-UGCGCCUGGAAUGGAA-----ACACAUGGCAAACAAAGCCCAAAGUUGGGGGG---------- (((.((((((((((((((((......)))))))))))))..))).-)))(((((...((...-----.)))).)))........((((....))))...---------- ( -31.10, z-score = -2.64, R) >droVir3.scaffold_12970 5123447 95 + 11907090 GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAA--UGCGCCUGGAACGUAGAAAGCAAGUUAUCUGGAACGGUAACGGGAACAGAU------------ ((((((((((((((((((((......)))))))))))))..)))--).)))(((...(((.....))..((((((......)))))).)...)))..------------ ( -27.90, z-score = -2.24, R) >droMoj3.scaffold_6473 4661596 101 + 16943266 GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAA--UGCGCCUGGAACGCACGAAGCUAGUUCUCUGGAACGGGAACGGGAACGGGAACAGAU------ (((.((((((((((((((((......)))))))))))).)))).--))).((((...(((.....))..((((((......)))))).)...)))).......------ ( -33.40, z-score = -2.49, R) >droGri2.scaffold_15081 4052509 84 - 4274704 GCGAUUCGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAA--UGCGCCUGGAACGU-----------UUUCUGGAACGGGAACGGGAACAGAU------------ ((((((((((((((((((((......)))))))))))))..)))--).)))(((...(((-----------(..(......)..))))....)))..------------ ( -32.10, z-score = -3.81, R) >consensus GCGAUUUGAAAAUUGUGUCGUCAGCUUGACACAAUUUUCCAGAAA_UGCGCCUGGAAUGGAGCGG__GGGAUAGCCAGACGAAAACCAGUAACAAAAACGGGAGCA___ (((.((((((((((((((((......)))))))))))))..)))....)))((((...................))))............................... (-19.09 = -18.88 + -0.21)
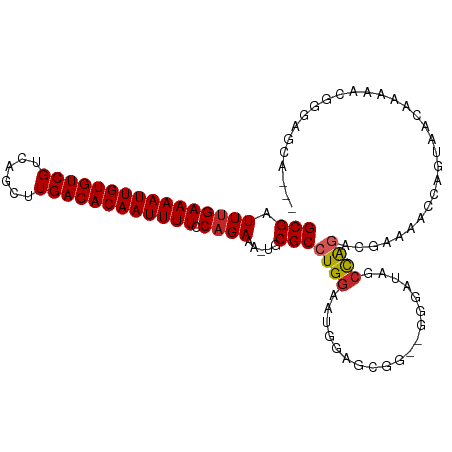
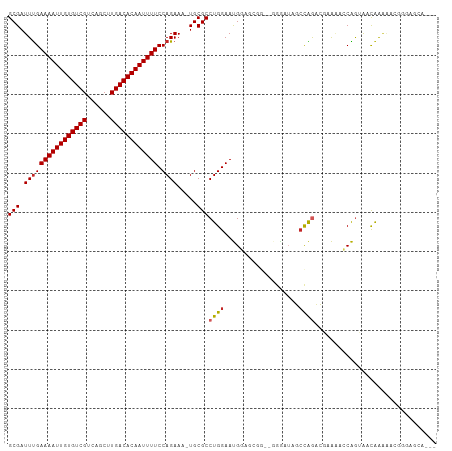
| Location | 20,798,267 – 20,798,375 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.89 |
| Shannon entropy | 0.53866 |
| G+C content | 0.46480 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20798267 108 - 22422827 CAGUGCUCCCGUUUUUGUUACUGGUUUUCGGCUGGCUAUUCC-UCCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC ..((((...((....))...((((.....(((.((.......-.))))).....))))))))........((((((((((((........))))))))))))....... ( -28.40, z-score = -1.55, R) >droEre2.scaffold_4690 17380236 107 - 18748788 CAUUGCUCCCGUUUUUGUUACUGGUUUUCGGCUGGCUAUCCC--CCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC ....((.........(((..((((.....(((.((......)--).))).....))))..))).......((((((((((((........)))))))))))).....)) ( -28.50, z-score = -1.84, R) >droYak2.chrX 20014882 107 - 21770863 CAUUGCUCCCGUUUUUGUUACUGGUUUUCGGCUGGCUAUCCC--CCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC ....((.........(((..((((.....(((.((......)--).))).....))))..))).......((((((((((((........)))))))))))).....)) ( -28.50, z-score = -1.84, R) >droSec1.super_8 3060483 108 - 3762037 CAUUGCUCCCGUUUUUGUUACUGGGUUUCGGCUGGCUAUCCC-UCCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC ....((.........(((..(((((....(((.((.......-.)))))....)))))..))).......((((((((((((........)))))))))))).....)) ( -28.10, z-score = -1.26, R) >droSim1.chrX 15966785 108 - 17042790 CAUUGCUCCCGUUUUUGUUACUGGGUUUCGGCUGGCUAUCCC-UCCGCUCCAUUCCAGGCGCAUUUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC ....((.........(((..(((((....(((.((.......-.)))))....)))))..))).......((((((((((((........)))))))))))).....)) ( -28.10, z-score = -1.26, R) >dp4.chrXL_group1e 9446010 99 - 12523060 ---CUCUACUUCUGUUGUUGUUGUUGUUUUUUUGGAUUUUGU-----GUUCAUUCCAGGCGCA--UUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC ---...........................((((((..((((-----(((..(((((((....--.)))))))......(((.....))))))))))..)))))).... ( -22.50, z-score = -1.09, R) >droAna3.scaffold_13334 135805 93 + 1562580 ----------CCCCCCAACUUUGGGCUUUGUUUGCCAUGUGU-----UUCCAUUCCAGGCGCA-UUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC ----------.............(((.......)))((((((-----((.......)))))))-).....((((((((((((........))))))))))))....... ( -25.00, z-score = -1.66, R) >droVir3.scaffold_12970 5123447 95 - 11907090 ------------AUCUGUUCCCGUUACCGUUCCAGAUAACUUGCUUUCUACGUUCCAGGCGCA--UUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC ------------(((((....((....))...))))).....................(((.(--((...((((((((((((........)))))))))))).)))))) ( -21.10, z-score = -1.45, R) >droMoj3.scaffold_6473 4661596 101 - 16943266 ------AUCUGUUCCCGUUCCCGUUCCCGUUCCAGAGAACUAGCUUCGUGCGUUCCAGGCGCA--UUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC ------................((((.(......).))))..((...((((((.....)))))--)....((((((((((((........)))))))))))).....)) ( -26.40, z-score = -1.82, R) >droGri2.scaffold_15081 4052509 84 + 4274704 ------------AUCUGUUCCCGUUCCCGUUCCAGAAA-----------ACGUUCCAGGCGCA--UUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCGAAUCGC ------------...............((((((((((.-----------.(((.....)))..--)))))((((((((((((........))))))))))))))).)). ( -24.10, z-score = -2.52, R) >consensus ___UGCUCCCGUUUUUGUUACUGGUUUUCGGCUGGCUAUCCC__CCGCUCCAUUCCAGGCGCA_UUUCUGGAAAAUUGUGUCAAGCUGACGACACAAUUUUCAAAUCGC .......................................................((((.......))))((((((((((((........))))))))))))....... (-17.30 = -17.30 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:11 2011