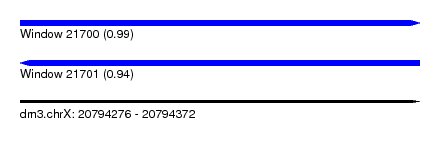
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,794,276 – 20,794,372 |
| Length | 96 |
| Max. P | 0.988218 |

| Location | 20,794,276 – 20,794,372 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 72.84 |
| Shannon entropy | 0.53459 |
| G+C content | 0.44413 |
| Mean single sequence MFE | -24.19 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
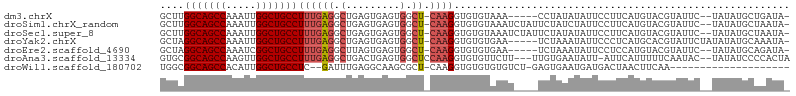
>dm3.chrX 20794276 96 + 22422827 -UAUCAGCAUAUA--GAAUACGUACAUGAAGGAAUAUAUAGG-----UUUACACACCUUG-AGCCACUCACUCAGCCUCAAAGGCAGCCAAUUUGGCUGCCAAGC -.....((.....--...........(((.((.......(((-----(......))))((-((.......)))).)))))..(((((((.....)))))))..)) ( -24.40, z-score = -2.13, R) >droSim1.chrX_random 412897 101 + 5698898 -UAUUAGCAUAUA--GAAUACGUACAUGAAGGAAUAGAUAGAAUAGAUUUACACACCUUG-AGCCACUCACUCAGCCUCAAAGGCAGCCAAUUUGGCUGCCAAGC -.....((.....--...........(((.((..(((((.......)))))......(((-((.......))))))))))..(((((((.....)))))))..)) ( -21.80, z-score = -1.60, R) >droSec1.super_8 3056345 101 + 3762037 -UAUUAGCAUAUA--GAAUACGUACAUGAAGGAAUAUAUAGAAUAGAUUUACACACCUUG-AGCCACUCACUCAGCCUCAAAGGCAGCCAAUUUGGCUGCCAAGC -.....((.....--...........(((.((......((((.....))))......(((-((.......))))))))))..(((((((.....)))))))..)) ( -20.70, z-score = -1.29, R) >droYak2.chrX 20010806 98 + 21770863 -UAUUUGCAUAUAUAGAAUACGUGCAUGAGGGAAUAUUUAGA-----UUCACACACCUUG-AGCCACUCACUCAGCCUCAAAGGCAGCCAAUUUGGCUGCCUAGC -....((((..(((...)))..))))(((((((((......)-----))).......(((-((.......)))))))))).((((((((.....))))))))... ( -31.60, z-score = -3.60, R) >droEre2.scaffold_4690 17376039 96 + 18748788 -UAUCUGCAUAUA--GAAUACGUACAUGGAGGAAUAUUUAGA-----UUCACACACCUUG-AGCCACUCACUAAGCCUCAAAGGCAGCCGAUUUGGCUGCCUAGC -.....((.....--.............((((....(((((.-----...........((-((...)))))))))))))..((((((((.....)))))))).)) ( -22.00, z-score = -0.88, R) >droAna3.scaffold_13334 132278 99 - 1562580 UAGUGGGGAUAUA--GUAUUGAAAAAUGAAU-AAUAUUCACAA---AAGAACACACCUUGGAGCCACUCAGUCAGCCUCAAAGGCAGCCAACUUGGCUGCCGCAC .(((((((((((.--.((((....))))...-.))))))....---(((.......)))....)))))..............(((((((.....))))))).... ( -26.20, z-score = -1.75, R) >droWil1.scaffold_180702 2023345 81 - 4511350 --------------------UUGAAGUUAGUCAUCAUUCACUC-AGACACACACACCUUG-AGCGCUUGCCUCAAAUC--GAGGCAGCCAAUGUGGCUGCCGCCA --------------------((((.....(((...........-.))).........(((-((.(....)))))).))--))(((((((.....))))))).... ( -22.60, z-score = -1.28, R) >consensus _UAUUAGCAUAUA__GAAUACGUACAUGAAGGAAUAUUUAGA____AUUCACACACCUUG_AGCCACUCACUCAGCCUCAAAGGCAGCCAAUUUGGCUGCCAAGC ..................................................................................(((((((.....))))))).... (-15.46 = -15.46 + 0.00)

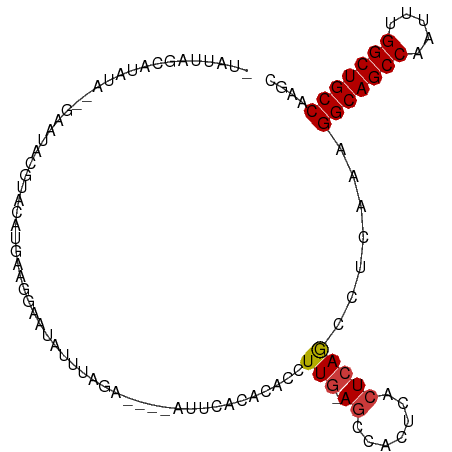
| Location | 20,794,276 – 20,794,372 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Shannon entropy | 0.53459 |
| G+C content | 0.44413 |
| Mean single sequence MFE | -28.29 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20794276 96 - 22422827 GCUUGGCAGCCAAAUUGGCUGCCUUUGAGGCUGAGUGAGUGGCU-CAAGGUGUGUAAA-----CCUAUAUAUUCCUUCAUGUACGUAUUC--UAUAUGCUGAUA- ....(((((((.....)))))))..(((((.(((((.....)))-))((((......)-----)))........))))).....((((..--...)))).....- ( -28.20, z-score = -1.20, R) >droSim1.chrX_random 412897 101 - 5698898 GCUUGGCAGCCAAAUUGGCUGCCUUUGAGGCUGAGUGAGUGGCU-CAAGGUGUGUAAAUCUAUUCUAUCUAUUCCUUCAUGUACGUAUUC--UAUAUGCUAAUA- ....(((((((.....)))))))..(((((..(((((..(((..-..((((......))))...)))..)))))))))).....((((..--...)))).....- ( -26.90, z-score = -1.03, R) >droSec1.super_8 3056345 101 - 3762037 GCUUGGCAGCCAAAUUGGCUGCCUUUGAGGCUGAGUGAGUGGCU-CAAGGUGUGUAAAUCUAUUCUAUAUAUUCCUUCAUGUACGUAUUC--UAUAUGCUAAUA- ....(((((((.....)))))))..(((((..(((((.((((..-..((((......))))...)))).)))))))))).....((((..--...)))).....- ( -26.80, z-score = -0.93, R) >droYak2.chrX 20010806 98 - 21770863 GCUAGGCAGCCAAAUUGGCUGCCUUUGAGGCUGAGUGAGUGGCU-CAAGGUGUGUGAA-----UCUAAAUAUUCCCUCAUGCACGUAUUCUAUAUAUGCAAAUA- ...((((((((.....)))))))).(((((.(((((.....)))-))((((......)-----)))........)))))((((..(((...)))..))))....- ( -32.00, z-score = -2.03, R) >droEre2.scaffold_4690 17376039 96 - 18748788 GCUAGGCAGCCAAAUCGGCUGCCUUUGAGGCUUAGUGAGUGGCU-CAAGGUGUGUGAA-----UCUAAAUAUUCCUCCAUGUACGUAUUC--UAUAUGCAGAUA- (((((((((((.....))))))))....)))...(((.((((..-..(((.((((...-----.....)))).))))))).)))((((..--...)))).....- ( -29.50, z-score = -1.51, R) >droAna3.scaffold_13334 132278 99 + 1562580 GUGCGGCAGCCAAGUUGGCUGCCUUUGAGGCUGACUGAGUGGCUCCAAGGUGUGUUCUU---UUGUGAAUAUU-AUUCAUUUUUCAAUAC--UAUAUCCCCACUA (((.(((((((.....)))))))((((..(((........)))..))))..(((((...---..((((((...-)))))).....)))))--........))).. ( -27.90, z-score = -1.25, R) >droWil1.scaffold_180702 2023345 81 + 4511350 UGGCGGCAGCCACAUUGGCUGCCUC--GAUUUGAGGCAAGCGCU-CAAGGUGUGUGUGUCU-GAGUGAAUGAUGACUAACUUCAA-------------------- (((((((((((.....)))))))((--(....((.(((.((((.-....)))).))).)).-...))).....).))).......-------------------- ( -26.70, z-score = -0.74, R) >consensus GCUUGGCAGCCAAAUUGGCUGCCUUUGAGGCUGAGUGAGUGGCU_CAAGGUGUGUAAAU____UCUAAAUAUUCCUUCAUGUACGUAUUC__UAUAUGCUAAUA_ ....(((((((.....)))))))((((.((((........)))).))))........................................................ (-18.46 = -18.93 + 0.47)
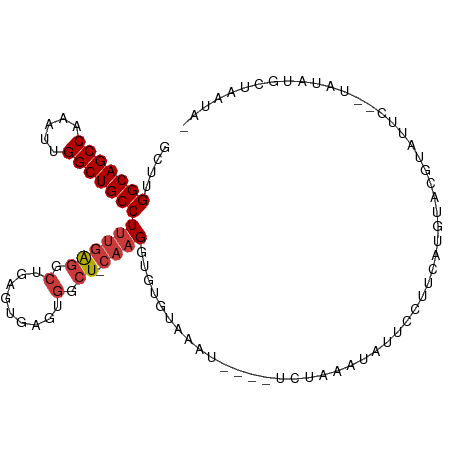
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:08 2011