
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,784,820 – 20,784,996 |
| Length | 176 |
| Max. P | 0.957928 |

| Location | 20,784,820 – 20,784,933 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.45 |
| Shannon entropy | 0.13497 |
| G+C content | 0.44620 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.957928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20784820 113 + 22422827 AACCAAAUGCAUUCCACUGCAACUG-AACUUCCUUUGAGCCCCAUCCACAUGUGGCAAUUAUAUGUGUGUGCUUCCCACGACCCUUCGCUCUU---UUUGGGGCUCAAAUUAAAUUU ...((..((((......))))..))-.......(((((((((((..((((((((.....))))))))((((.....)))).............---..)))))))))))........ ( -30.60, z-score = -3.43, R) >droYak2.chrX 20000946 116 + 21770863 AACCAAAUGCAUUCCACUGCAACAGCAACUUCCUUUGAGCCCUUUCCACAUGUGGCAAUUAUUUGUGUGUGCGGCCU-CGCCUUUCCACUCUUCUUUUUGGGGCUCAAAUUAAAUUU .......((((......))))............((((((((((...(((((...((((....))))))))).(((..-.))).................))))))))))........ ( -26.80, z-score = -1.30, R) >droSec1.super_8 3047091 113 + 3762037 AACCAAAUGCAUUCCACUGCAACUG-AACUUCCUUUGAGCCCCUUCCACAUGUGGCAAUUAUUUGUGUGUGCUUCCCACGCCCCACCGCUCUU---UUUGGGGCUCAAAUUAAAUUU ...((..((((......))))..))-.......((((((((((........(((((((....))).(((((.....)))))....))))....---...))))))))))........ ( -30.16, z-score = -3.19, R) >droSim1.chrX_random 403659 113 + 5698898 AACCAAAUGCAUUCCACUGCAACUG-AACUUCCUUUGAGCCCUUUCCACAUGUGGCAAUUAUUUGUGUGUGCUUCCCAUGCCCCACCGCUCUU---UUUGGGGCUCAAAUUAAAUUU ...((..((((......))))..))-.......((((((((((........(((((((....))))(((.((.......))..))))))....---...))))))))))........ ( -26.26, z-score = -1.70, R) >consensus AACCAAAUGCAUUCCACUGCAACUG_AACUUCCUUUGAGCCCCUUCCACAUGUGGCAAUUAUUUGUGUGUGCUUCCCACGCCCCACCGCUCUU___UUUGGGGCUCAAAUUAAAUUU .......((((......))))............((((((((((........(((((((....))))(((((.....))))).....)))..........))))))))))........ (-24.58 = -24.52 + -0.06)



| Location | 20,784,820 – 20,784,933 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.45 |
| Shannon entropy | 0.13497 |
| G+C content | 0.44620 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -26.47 |
| Energy contribution | -27.04 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.539969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20784820 113 - 22422827 AAAUUUAAUUUGAGCCCCAAA---AAGAGCGAAGGGUCGUGGGAAGCACACACAUAUAAUUGCCACAUGUGGAUGGGGCUCAAAGGAAGUU-CAGUUGCAGUGGAAUGCAUUUGGUU .(((((..(((((((((((..---..((.(....).))((((.((..............)).)))).......)))))))))))..)))))-(((.((((......)))).)))... ( -38.24, z-score = -3.31, R) >droYak2.chrX 20000946 116 - 21770863 AAAUUUAAUUUGAGCCCCAAAAAGAAGAGUGGAAAGGCG-AGGCCGCACACACAAAUAAUUGCCACAUGUGGAAAGGGCUCAAAGGAAGUUGCUGUUGCAGUGGAAUGCAUUUGGUU .(((((..(((((((((...........(((.....(((-....)))...))).........(((....)))...)))))))))..)))))((((.((((......))))..)))). ( -32.70, z-score = -1.23, R) >droSec1.super_8 3047091 113 - 3762037 AAAUUUAAUUUGAGCCCCAAA---AAGAGCGGUGGGGCGUGGGAAGCACACACAAAUAAUUGCCACAUGUGGAAGGGGCUCAAAGGAAGUU-CAGUUGCAGUGGAAUGCAUUUGGUU .(((((..((((((((((...---.......(((..((.......))...))).........(((....)))..))))))))))..)))))-(((.((((......)))).)))... ( -36.10, z-score = -2.28, R) >droSim1.chrX_random 403659 113 - 5698898 AAAUUUAAUUUGAGCCCCAAA---AAGAGCGGUGGGGCAUGGGAAGCACACACAAAUAAUUGCCACAUGUGGAAAGGGCUCAAAGGAAGUU-CAGUUGCAGUGGAAUGCAUUUGGUU .(((((..(((((((((....---.......(((..((.......))...))).........(((....)))...)))))))))..)))))-(((.((((......)))).)))... ( -31.20, z-score = -0.94, R) >consensus AAAUUUAAUUUGAGCCCCAAA___AAGAGCGGAGGGGCGUGGGAAGCACACACAAAUAAUUGCCACAUGUGGAAAGGGCUCAAAGGAAGUU_CAGUUGCAGUGGAAUGCAUUUGGUU .(((((..((((((((((..........((...............))...............(((....)))..))))))))))..))))).(((.((((......)))).)))... (-26.47 = -27.04 + 0.56)


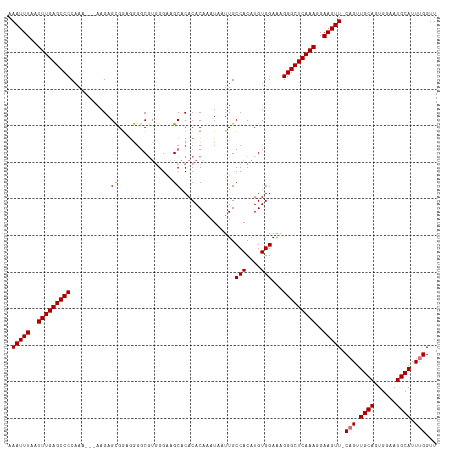
| Location | 20,784,880 – 20,784,996 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.09 |
| Shannon entropy | 0.20928 |
| G+C content | 0.40472 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -27.40 |
| Energy contribution | -28.15 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.632781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20784880 116 - 22422827 -CAUUUUUCUUUGUCCCGAUAUCAGCAUUUGGUUGAACUUUUUGCAUUUGGCCAAACAAAUUGUAAAUUUAAUUUGAGCCCCAAAAAGAGCGAAGGGUCGUGGGAAGCACACACAUA-- -...((((((.((.(((....(((((.....))))).(((((((.....(((....(((((((......))))))).))).)))))))......))).)).))))))..........-- ( -26.30, z-score = -0.49, R) >droYak2.chrX 20001007 119 - 21770863 CAAUUUUUCUUUGUCCCGAUAUCAGCAUUCUGCUGAACUUUUUGGCUUUGGCCAAACAAAUUGUAAAUUUAAUUUGAGCCCCAAAAAGAAGAGUGGAAAGGCGAGGCCGCACACACAAA .......((((..........(((((.....))))).((((((((....(((....(((((((......))))))).)))))))))))))))(((.....(((....)))...)))... ( -30.00, z-score = -1.22, R) >droSec1.super_8 3047151 116 - 3762037 -CAUUUCUCUUUGUCCCGAUAUCAGCAUUUUGUUGAACUGUUUGGCUUUGGCCAAACAAAUUGUAAAUUUAAUUUGAGCCCCAAAAAGAGCGGUGGGGCGUGGGAAGCACACACAAA-- -..((((((..((((((....(((((.....)))))((((((((((....)))...(((((((......)))))))...........))))))))))))).))))))..........-- ( -34.60, z-score = -1.63, R) >droSim1.chrX_random 403719 116 - 5698898 -CAUUUUUCUUUGUCCCGGUAUCAGCAUUUUGUUGAACUUUUUGGCUUUGGCCAAACAAAUUGUAAAUUUAAUUUGAGCCCCAAAAAGAGCGGUGGGGCAUGGGAAGCACACACAAA-- -...((((((.(((((((.(.(((((.....))))).((((((((....(((....(((((((......))))))).)))))))))))...).))))))).))))))..........-- ( -34.80, z-score = -1.81, R) >consensus _CAUUUUUCUUUGUCCCGAUAUCAGCAUUUUGUUGAACUUUUUGGCUUUGGCCAAACAAAUUGUAAAUUUAAUUUGAGCCCCAAAAAGAGCGGUGGGGCGUGGGAAGCACACACAAA__ .....(((((.((((((....(((((.....))))).((((((((....(((....(((((((......))))))).)))))))))))......)))))).)))))............. (-27.40 = -28.15 + 0.75)

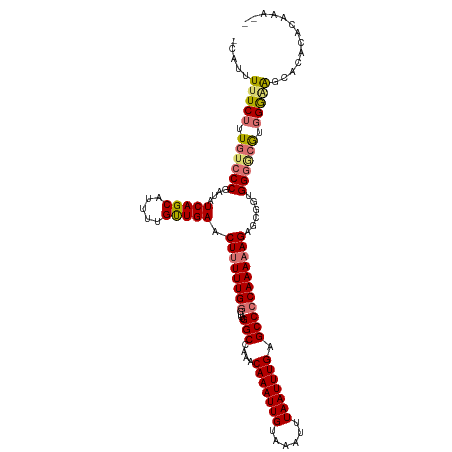
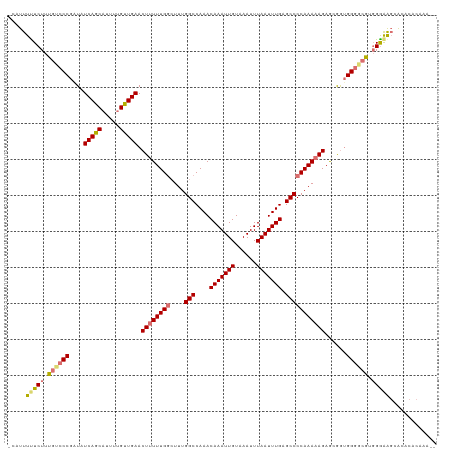
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:06 2011