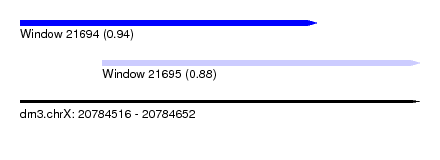
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,784,516 – 20,784,652 |
| Length | 136 |
| Max. P | 0.943373 |

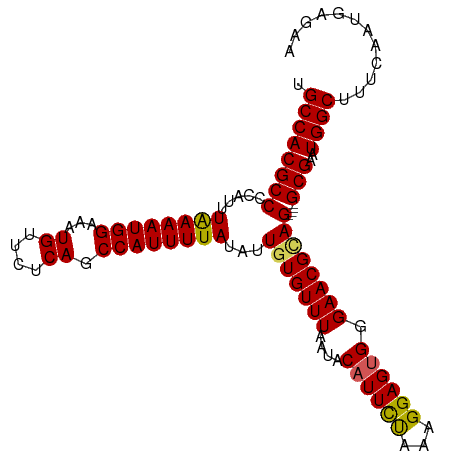
| Location | 20,784,516 – 20,784,617 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.22 |
| Shannon entropy | 0.08205 |
| G+C content | 0.39493 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -25.24 |
| Energy contribution | -24.88 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20784516 101 + 22422827 UGCCACGCCCCAUUUGAAAUGGAAAUGUUCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUCUAAAGGAGUGGGAACGCAG----GCGAAUGGCUUUCAAUGAGAA .((((((((.....((((((((...((....)).))))))))...(((((((....(((((.....))))).))))))))----)))..))))............ ( -31.70, z-score = -2.85, R) >droEre2.scaffold_4690 17366167 105 + 18748788 UGCCACGCCCCAUUUAAAAUGGAAAUGUUCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUUUAAAGGAGUGGGAACGAAGGCGGGCGAAUGGCUUUCAAUGAAAA .(((.((((((((.....))))...((((((((.((..((((...(((((...)))))...))))))..))))))))..)))))))(((.....)))........ ( -30.00, z-score = -2.05, R) >droYak2.chrX 20000640 101 + 21770863 UGCCACGCCCCAUUUAAAAUGGAAAUGUUCUCAGCCAUUUUAUAUUGUGUUUAAUACUUUUUAAAGGAGUGGGAACGUAG----GCGAAUGGCUUUCAAUGAAAA .((((((((((((.....))))..(((((((((.((..((((.(..((((...))))..).))))))..))))))))).)----)))..))))............ ( -27.60, z-score = -2.28, R) >droSec1.super_8 3046787 101 + 3762037 UGCCACGCCCCAUUUAAAAUGGAAAUGUUCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUCCAAAGGAGUGGGAACGCAG----GCGAAUGGCUUUCAAUGAGAA .((((((((.....((((((((...((....)).))))))))...(((((((....((((((...)))))).))))))))----)))..))))............ ( -33.00, z-score = -3.32, R) >droSim1.chrX_random 403355 101 + 5698898 UGCCACGCCCCAUUUAAAAUGGAAAUGUUCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUCCAAAGGAGUGGGAACGCAG----GCGAAUGGCUUUCAAUGAGAA .((((((((.....((((((((...((....)).))))))))...(((((((....((((((...)))))).))))))))----)))..))))............ ( -33.00, z-score = -3.32, R) >consensus UGCCACGCCCCAUUUAAAAUGGAAAUGUUCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUCUAAAGGAGUGGGAACGCAG____GCGAAUGGCUUUCAAUGAGAA .(((((((......((((((((...((....)).))))))))..((((((((....((((((...)))))).))))))))....)))..))))............ (-25.24 = -24.88 + -0.36)

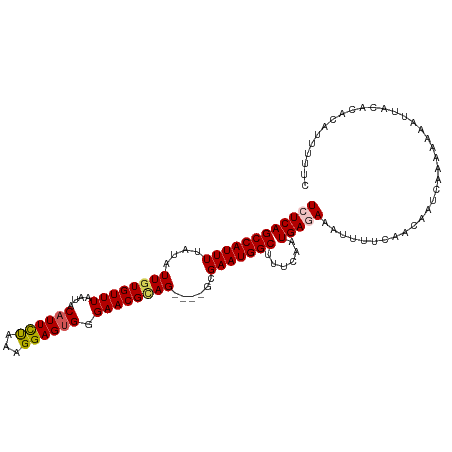
| Location | 20,784,544 – 20,784,652 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.80 |
| Shannon entropy | 0.08981 |
| G+C content | 0.31978 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20784544 108 + 22422827 UCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUCUAAAGGAGUGGGAACGCAG----GCGAAUGGCUUUCAAUGAGAAAUUUUCAACAAUCAAAAAAAUUACACACAUUUUUC ((((((((((((....((((((((....(((((.....))))).))))))))----..)))))))......))))).................................... ( -25.10, z-score = -3.35, R) >droEre2.scaffold_4690 17366195 112 + 18748788 UCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUUUAAAGGAGUGGGAACGAAGGCGGGCGAAUGGCUUUCAAUGAAAAAUUUGCAACAAUCAAAAAAAUUACACACAUUUUUC (((((.((..((((...(((((...)))))...))))))..)))))..((((((.(.....).))))))...(((((((.((....................)).))))))) ( -17.45, z-score = 0.04, R) >droYak2.chrX 20000668 108 + 21770863 UCUCAGCCAUUUUAUAUUGUGUUUAAUACUUUUUAAAGGAGUGGGAACGUAG----GCGAAUGGCUUUCAAUGAAAAAUUUUCAACAAUCAAAAAAAUUACACACAUUUUUC ....((((((((....((((((((..((((((.....)))))).))))))))----..)))))))).....((((.....))))............................ ( -16.50, z-score = -0.54, R) >droSec1.super_8 3046815 108 + 3762037 UCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUCCAAAGGAGUGGGAACGCAG----GCGAAUGGCUUUCAAUGAGAAAUUUUUAAAAAUCAAAAAAAUUACACACAUUUUUC ((((((((((((....((((((((....((((((...)))))).))))))))----..)))))))......)))))(((((((.........)))))))............. ( -26.70, z-score = -3.63, R) >droSim1.chrX_random 403383 108 + 5698898 UCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUCCAAAGGAGUGGGAACGCAG----GCGAAUGGCUUUCAAUGAGAAAUUUUCAACAAUCAAAAAAAUUACACACAUUUUUC ((((((((((((....((((((((....((((((...)))))).))))))))----..)))))))......))))).................................... ( -26.50, z-score = -3.60, R) >consensus UCUCAGCCAUUUUAUAUUGUGUUUAAUACAUUCUAAAGGAGUGGGAACGCAG____GCGAAUGGCUUUCAAUGAGAAAUUUUCAACAAUCAAAAAAAUUACACACAUUUUUC ((((((((((((....((((((((....((((((...)))))).))))))))......)))))))......))))).................................... (-18.98 = -19.18 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:04 2011