| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,776,066 – 20,776,161 |
| Length | 95 |
| Max. P | 0.662568 |

| Location | 20,776,066 – 20,776,161 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 61.12 |
| Shannon entropy | 0.64416 |
| G+C content | 0.51635 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -9.72 |
| Energy contribution | -11.56 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
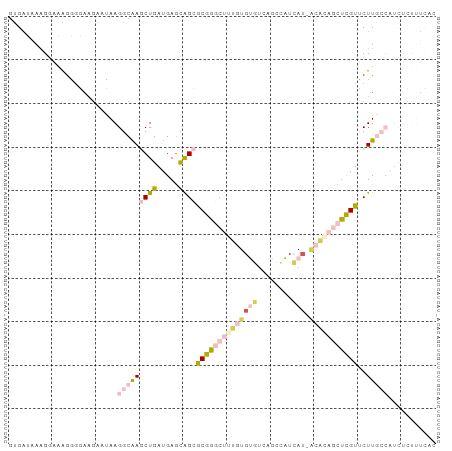
>dm3.chrX 20776066 95 + 22422827 GUGAUAAGGGAAAUGUGAAGAAUAAGGCAAGCUGAUGAACAGCGCGGGCUUUGUGUGUCAGCCAUCA---CAGAGCUCGUUCUUGCCAUCUCUUUCAC ((((.((((((..(((.....))).(((((((((.....))))((((((((((((((.....)).))---))))))))))..))))).)))))))))) ( -38.10, z-score = -3.87, R) >droSec1.super_8 3038457 98 + 3762037 GUGAUAAAGGAAAGAGGAAGCAUAAGGCAAGCUGAUGAGCAGCACGGGCUUUGUGUGUCAGCCAUCAUCACACAGCUCGUUCUUGCCAUCUCUUUCAC .........((((((((........(((((((((.....))))(((((((.((((((........)).)))).)))))))..))))).)))))))).. ( -33.30, z-score = -2.12, R) >droSim1.chrX 15957491 98 + 17042790 GUGAUAAAGGAAAGGGGAAGCAUAAGGCAAGCUGAUGAGCAGCGCGGGCUUUGUGUGUCAGCCAUCAUCACACAGCUCGUUCUUGCCAUCUCUUUCAC .........((((((((........(((((((((.....))))(((((((.((((((........)).)))).)))))))..))))).)))))))).. ( -32.70, z-score = -1.54, R) >droVir3.scaffold_12970 6810793 82 - 11907090 GCGAGUGUGAGUGGCGCGCGGCGAAGGCGA-CUGAUGAGUGGGGCAAACUGCUUGGCUGCUGUUUUAU-GUUCCACUUGUGCUU-------------- ..(((((..(((((.(((((((..((....-))....(((.((((.....)))).)))))))).....-)).)))))..)))))-------------- ( -29.70, z-score = -0.78, R) >droGri2.scaffold_15081 246915 82 + 4274704 GUGGGUUGGGGUGGGGCAAGACGAAGGCAA-CUGAUGAGUGGGACAAACUGCUUGGGUCCUGUUUUAU-GUGCCACUUGUACUU-------------- ..((((..((((((.(((......((....-))...(((..((((...(.....).))))..)))..)-)).))))))..))))-------------- ( -24.50, z-score = -0.63, R) >consensus GUGAUAAAGGAAAGGGGAAGAAUAAGGCAAGCUGAUGAGCAGCGCGGGCUUUGUGUGUCAGCCAUCAU_ACACAGCUCGUUCUUGCCAUCUCUUUCAC .........................(((((((((.....))))(((((((((((...............)))))))))))..)))))........... ( -9.72 = -11.56 + 1.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:05:02 2011