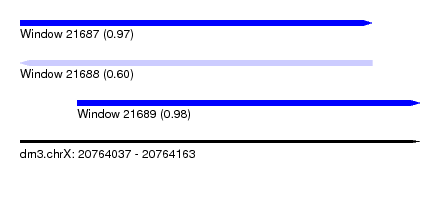
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,764,037 – 20,764,163 |
| Length | 126 |
| Max. P | 0.984218 |

| Location | 20,764,037 – 20,764,148 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Shannon entropy | 0.39824 |
| G+C content | 0.40150 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.15 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
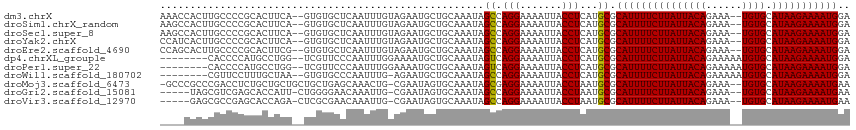
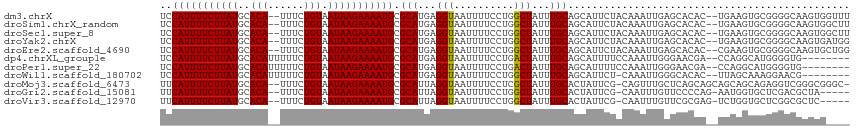
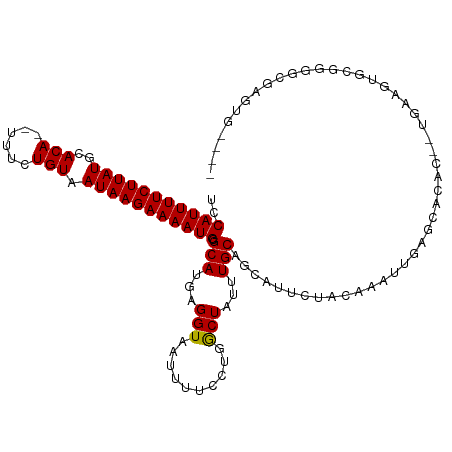
>dm3.chrX 20764037 111 + 22422827 AAACCACUUGCCCCGCACUUCA--GUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGA ...(((..(((...((((....--..))))...((((((((....)))))))).((.(((.......)))...)))))(((((((((((((....--)))).)))))))))))). ( -30.90, z-score = -2.96, R) >droSim1.chrX_random 5373122 111 + 5698898 AAGCCACUUGCCCCGCACUUCA--GUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGA .((((((((((...)))....)--))).)))..((((((((....)))))))).((.(((.......)))...)).(((((((((((((((....--)))).))))))))))).. ( -31.30, z-score = -2.52, R) >droSec1.super_8 3026145 111 + 3762037 AAGCCACUUGCCCCGCACUUCA--GUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGA .((((((((((...)))....)--))).)))..((((((((....)))))))).((.(((.......)))...)).(((((((((((((((....--)))).))))))))))).. ( -31.30, z-score = -2.52, R) >droYak2.chrX 19980017 111 + 21770863 CCAUCACUUGCCCCGCACUUCA--GUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGA ...((.((.((...((((....--..))))...((((((((....)))))))).)).))))...............(((((((((((((((....--)))).))))))))))).. ( -31.40, z-score = -2.87, R) >droEre2.scaffold_4690 17346162 111 + 18748788 CCAGCACUUGCCCCGCACUUCG--GUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGA ...(((((.((..(((((....--)))))....((((((((....)))))))).)).))((........)).))).(((((((((((((((....--)))).))))))))))).. ( -35.10, z-score = -3.31, R) >dp4.chrXL_group1e 9409727 105 + 12523060 --------CACCCCAUGCCUGG--UCGUUCCCAAUUUGGAAAAUGCUGCAAAUAGUCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAAAAUGUGCAUAAGAAAAUGGA --------....((((.(((((--..(((..((..((....))...))..)))..)))))........((.(((((((((((((......)).))))))))))).))...)))). ( -25.60, z-score = -1.80, R) >droPer1.super_22 892274 105 - 1688296 --------CACCCCAUGCCUGG--UCGUUCCCAAUUUGGAAAAUGCUGCAAAUAGUCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAAAAUGUGCAUAAGAAAAUGGA --------....((((.(((((--..(((..((..((....))...))..)))..)))))........((.(((((((((((((......)).))))))))))).))...)))). ( -25.60, z-score = -1.80, R) >droWil1.scaffold_180702 2518543 104 + 4511350 --------CGUUCCUUUGCUAA--GUGUGCCCAAUUUG-AGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAAAAUGUGCAUAAGAAAAUGGA --------..(((((..((((.--.(((((........-.....)).)))..)))).)))))..............(((((((((((((((......)))).))))))))))).. ( -24.82, z-score = -1.66, R) >droMoj3.scaffold_6473 4608426 111 + 16943266 -GCCCGCCCGACCUCUGCUGCUGCUGCUGAGCAAACUG-CGAAUAGUGCAAAUAGCGAGGAAAAUUACCUAAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGAA -...(((....((((.((((.(((.((((.((.....)-)...)))))))..))))))))...(((....))))))(((((((((((((((....--)))).))))))))))).. ( -32.10, z-score = -3.20, R) >droGri2.scaffold_15081 4016468 106 - 4274704 -----UAGCGUCGAGCACCAUU-CUGGGGAACAAAUUG-CGAAUAGUGCAAAUAGCCAGGAAAAUUACCUAAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGAA -----..((((...((((.(((-(.(.((......)).-))))).))))........(((.......))).)))).(((((((((((((((....--)))).))))))))))).. ( -27.20, z-score = -2.41, R) >droVir3.scaffold_12970 5083505 106 + 11907090 -----GAGCGCCGAGCACCAGA-CUCGCGAACAAAUUG-CGAAUAGUGCAAAUAGCCAGGAAAAUUACCUAAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGAA -----..((((...((((...(-.((((((.....)))-))).).))))........(((.......)))...))))((((((((((((((....--)))).))))))))))... ( -32.00, z-score = -4.60, R) >consensus ____CACUCGCCCCGCACUUCA__GUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA__UGUGCAUAAGAAAAUGGA ......................................................((.(((.......)))...)).(((((((((((((((......)))).))))))))))).. (-15.30 = -15.15 + -0.15)
| Location | 20,764,037 – 20,764,148 |
|---|---|
| Length | 111 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.03 |
| Shannon entropy | 0.39824 |
| G+C content | 0.40150 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -14.79 |
| Energy contribution | -14.65 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.604836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20764037 111 - 22422827 UCCAUUUUCUUAUGCACA--UUUCUGUAAUAAGAAAAUGCGCAUGAGGUAAUUUUCCUGGCUAUUUGCAGCAUUCUACAAAUUGAGCACAC--UGAAGUGCGGGGCAAGUGGUUU .......((((((((.((--(((((......)).))))).))))))))..........((((((((((..(..((........))((((..--....)))))..)))))))))). ( -31.60, z-score = -1.79, R) >droSim1.chrX_random 5373122 111 - 5698898 UCCAUUUUCUUAUGCACA--UUUCUGUAAUAAGAAAAUGCGCAUGAGGUAAUUUUCCUGGCUAUUUGCAGCAUUCUACAAAUUGAGCACAC--UGAAGUGCGGGGCAAGUGGCUU .......((((((((.((--(((((......)).))))).))))))))..........((((((((((..(..((........))((((..--....)))))..)))))))))). ( -34.10, z-score = -2.33, R) >droSec1.super_8 3026145 111 - 3762037 UCCAUUUUCUUAUGCACA--UUUCUGUAAUAAGAAAAUGCGCAUGAGGUAAUUUUCCUGGCUAUUUGCAGCAUUCUACAAAUUGAGCACAC--UGAAGUGCGGGGCAAGUGGCUU .......((((((((.((--(((((......)).))))).))))))))..........((((((((((..(..((........))((((..--....)))))..)))))))))). ( -34.10, z-score = -2.33, R) >droYak2.chrX 19980017 111 - 21770863 UCCAUUUUCUUAUGCACA--UUUCUGUAAUAAGAAAAUGCGCAUGAGGUAAUUUUCCUGGCUAUUUGCAGCAUUCUACAAAUUGAGCACAC--UGAAGUGCGGGGCAAGUGAUGG .(((((.((((((((.((--(((((......)).))))).))))))))..((((.(((.((.((((.(((..(((........)))....)--)))))))).))).))))))))) ( -29.80, z-score = -1.17, R) >droEre2.scaffold_4690 17346162 111 - 18748788 UCCAUUUUCUUAUGCACA--UUUCUGUAAUAAGAAAAUGCGCAUGAGGUAAUUUUCCUGGCUAUUUGCAGCAUUCUACAAAUUGAGCACAC--CGAAGUGCGGGGCAAGUGCUGG .......((((((((.((--(((((......)).))))).))))))))........(..((.((((((..(..((........))((((..--....)))))..))))))))..) ( -31.90, z-score = -1.57, R) >dp4.chrXL_group1e 9409727 105 - 12523060 UCCAUUUUCUUAUGCACAUUUUUCUGUAAUAAGAAAAUGCGCAUGAGGUAAUUUUCCUGACUAUUUGCAGCAUUUUCCAAAUUGGGAACGA--CCAGGCAUGGGGUG-------- .......((((((((.................((((((((((((.(((.......))).).....))).))))))))....((((......--))))))))))))..-------- ( -25.90, z-score = -1.14, R) >droPer1.super_22 892274 105 + 1688296 UCCAUUUUCUUAUGCACAUUUUUCUGUAAUAAGAAAAUGCGCAUGAGGUAAUUUUCCUGACUAUUUGCAGCAUUUUCCAAAUUGGGAACGA--CCAGGCAUGGGGUG-------- .......((((((((.................((((((((((((.(((.......))).).....))).))))))))....((((......--))))))))))))..-------- ( -25.90, z-score = -1.14, R) >droWil1.scaffold_180702 2518543 104 - 4511350 UCCAUUUUCUUAUGCACAUUUUUCUGUAAUAAGAAAAUGCGCAUGAGGUAAUUUUCCUGGCUAUUUGCAGCAUUCU-CAAAUUGGGCACAC--UUAGCAAAGGAACG-------- .......((((((((.((((((.((......)))))))).)))))))).....(((((.((((..(((..((....-.....)).)))...--.))))..)))))..-------- ( -26.80, z-score = -2.49, R) >droMoj3.scaffold_6473 4608426 111 - 16943266 UUCAUUUUCUUAUGCACA--UUUCUGUAAUAAGAAAAUGCGCAUUAGGUAAUUUUCCUCGCUAUUUGCACUAUUCG-CAGUUUGCUCAGCAGCAGCAGCAGAGGUCGGGCGGGC- ..(((((((((((..(((--....))).))))))))))).................((((((..((((.......)-)))((((((..((....)))))))).....)))))).- ( -27.30, z-score = -0.34, R) >droGri2.scaffold_15081 4016468 106 + 4274704 UUCAUUUUCUUAUGCACA--UUUCUGUAAUAAGAAAAUGCGCAUUAGGUAAUUUUCCUGGCUAUUUGCACUAUUCG-CAAUUUGUUCCCCAG-AAUGGUGCUCGACGCUA----- ..(((((((((((..(((--....))).))))))))))).((.(((((.......)))))......(((((((((.-..............)-)))))))).....))..----- ( -27.86, z-score = -2.98, R) >droVir3.scaffold_12970 5083505 106 - 11907090 UUCAUUUUCUUAUGCACA--UUUCUGUAAUAAGAAAAUGCGCAUUAGGUAAUUUUCCUGGCUAUUUGCACUAUUCG-CAAUUUGUUCGCGAG-UCUGGUGCUCGGCGCUC----- ...((((((((((..(((--....))).))))))))))((((.(((((.......)))))......((((((((((-(.........)))))-)..)))))...))))..----- ( -31.80, z-score = -3.51, R) >consensus UCCAUUUUCUUAUGCACA__UUUCUGUAAUAAGAAAAUGCGCAUGAGGUAAUUUUCCUGGCUAUUUGCAGCAUUCUACAAAUUGAGCACAC__UGAAGUGCGGGGCGAGUG____ ..(((((((((((..(((......))).))))))))))).(((...(((..........)))...)))............................................... (-14.79 = -14.65 + -0.15)
| Location | 20,764,055 – 20,764,163 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.47 |
| Shannon entropy | 0.26229 |
| G+C content | 0.34305 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.47 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
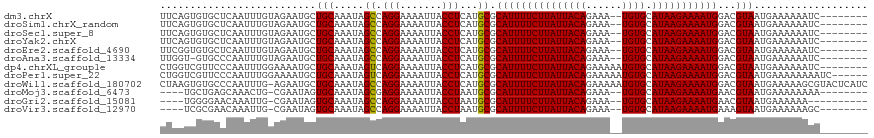
>dm3.chrX 20764055 108 + 22422827 UUCAGUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGACGUAAUGAAAAAAUC-------- (((..((.(((..((((((((....))))))))))))).))).......((((((((((((((((((((((....--)))).)))))))))))..))).)))).......-------- ( -30.90, z-score = -3.91, R) >droSim1.chrX_random 5373140 108 + 5698898 UUCAGUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGACGUAAUGAAAAAAUC-------- (((..((.(((..((((((((....))))))))))))).))).......((((((((((((((((((((((....--)))).)))))))))))..))).)))).......-------- ( -30.90, z-score = -3.91, R) >droSec1.super_8 3026163 108 + 3762037 UUCAGUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGACGUAAUGAAAAAAUC-------- (((..((.(((..((((((((....))))))))))))).))).......((((((((((((((((((((((....--)))).)))))))))))..))).)))).......-------- ( -30.90, z-score = -3.91, R) >droYak2.chrX 19980035 108 + 21770863 UUCAGUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGACGUAAUGAAAAAAUC-------- (((..((.(((..((((((((....))))))))))))).))).......((((((((((((((((((((((....--)))).)))))))))))..))).)))).......-------- ( -30.90, z-score = -3.91, R) >droEre2.scaffold_4690 17346180 108 + 18748788 UUCGGUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGACGUAAUGAAAAAAUC-------- ...((((.(((..((((((((....))))))))))).........))))((((((((((((((((((((((....--)))).)))))))))))..))).)))).......-------- ( -29.80, z-score = -3.38, R) >droAna3.scaffold_13334 93159 107 - 1562580 UUGGU-GUGCCCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGGACGUAAUGAAAAAAUC-------- ..(((-(..(((.((((((((....)))))))).)...)).....))))((((((((((((((((((((((....--)))).)))))))))))..))).)))).......-------- ( -29.70, z-score = -3.46, R) >dp4.chrXL_group1e 9409737 110 + 12523060 CUGGUCGUUCCCAAUUUGGAAAAUGCUGCAAAUAGUCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAAAAUGUGCAUAAGAAAAUGGACGUAAUGAAAAAAUC-------- ..(((..(((((.(((((.(......).))))).)...))))....)))((((((((((((((((((((((......)))).)))))))))))..))).)))).......-------- ( -24.30, z-score = -2.01, R) >droPer1.super_22 892284 112 - 1688296 CUGGUCGUUCCCAAUUUGGAAAAUGCUGCAAAUAGUCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAAAAUGUGCAUAAGAAAAUGGACGUAAUGAAAAAAAAUC------ ..(((..(((((.(((((.(......).))))).)...))))....)))((((((((((((((((((((((......)))).)))))))))))..))).)))).........------ ( -24.30, z-score = -2.09, R) >droWil1.scaffold_180702 2518553 117 + 4511350 CUAAGUGUGCCCAAUUUG-AGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAAAAUGUGCAUAAGAAAAUGGACGUAAUGAAAAAGCGUACUCAUC ................((-((.(((((((.....)).............((((((((((((((((((((((......)))).)))))))))))..))).))))...))))).)))).. ( -27.60, z-score = -2.27, R) >droMoj3.scaffold_6473 4608449 103 + 16943266 ----UGCUGAGCAAACUG-CGAAUAGUGCAAAUAGCGAGGAAAAUUACCUAAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGAACGUAAUGAAAAAAAA-------- ----.((((.....((((-....)))).....)))).(((.......)))..(((((((((((((((((((....--)))).)))))))))))..))))...........-------- ( -24.10, z-score = -3.04, R) >droGri2.scaffold_15081 4016486 101 - 4274704 ----UGGGGAACAAAUUG-CGAAUAGUGCAAAUAGCCAGGAAAAUUACCUAAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGAACGUAAUGAAAAAA---------- ----..........((((-((....(((((.......(((.......)))..)))))((((((((((((((....--)))).))))))))))...)))))).......---------- ( -23.40, z-score = -3.12, R) >droVir3.scaffold_12970 5083523 103 + 11907090 ----UCGCGAACAAAUUG-CGAAUAGUGCAAAUAGCCAGGAAAAUUACCUAAUGCGCAUUUUCUUAUUACAGAAA--UGUGCAUAAGAAAAUGAAAGUAAUGAAAAAAGC-------- ----((((((.....)))-)))...(.((.....)))(((.......)))..(((.(((((((((((((((....--)))).)))))))))))...)))...........-------- ( -25.30, z-score = -3.75, R) >consensus UUCAGUGUGCUCAAUUUGUAGAAUGCUGCAAAUAGCCAGGAAAAUUACCUCAUGCGCAUUUUCUUAUUACAGAAA__UGUGCAUAAGAAAAUGGACGUAAUGAAAAAAUC________ ..........................(((.....((.(((.......)))...)).(((((((((((((((......)))).)))))))))))...)))................... (-16.60 = -16.47 + -0.14)
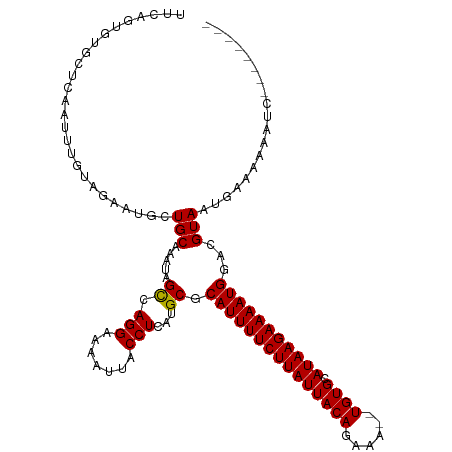
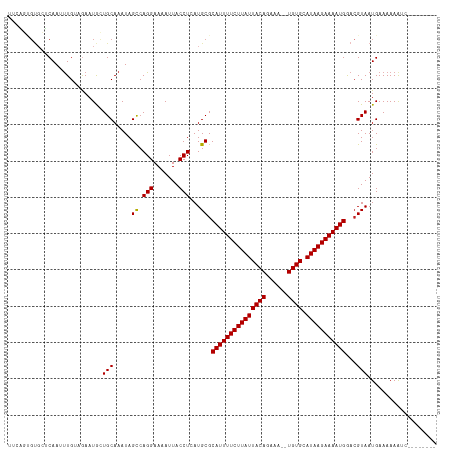
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:59 2011