| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,191,464 – 12,191,598 |
| Length | 134 |
| Max. P | 0.983893 |

| Location | 12,191,464 – 12,191,566 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 56.02 |
| Shannon entropy | 0.91502 |
| G+C content | 0.41488 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -10.49 |
| Energy contribution | -10.71 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.89 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
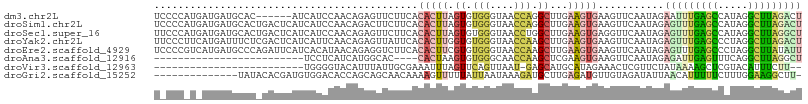
>dm3.chr2L 12191464 102 + 23011544 UCCCCAUGAUGAUGCAC------AUCAUCCAACAGAGUUCUUCACACUUAGUGUGGGUAACCAGGCUUGAAGUGAAGUUCAAUAGAAUUUGAGCCAUAGGCUUAGACU .......((((((....------)))))).......(..((((((....(((.(((....))).)))....))))))..).......((((((((...)))))))).. ( -30.10, z-score = -2.17, R) >droSim1.chr2L 11992293 108 + 22036055 UCCCCAUGAUGAUGCACUGACUCAUCAUCCAACAGACUUCUUCACACUUAGUGUGGGUAACCAGGCUUGAAGUGAAGUUCAAUAGAGUUUGAGCCAUAGGCUUAGACU .......(((((((........))))))).....((...((((((....(((.(((....))).)))....)))))).)).....((((((((((...)))))))))) ( -34.60, z-score = -2.54, R) >droSec1.super_16 386938 108 + 1878335 UUCCCAUGAUGAUGCACUGACUCAUCAUCCAACAGAGUUCUUCACACUUAGUGUGGGUAACCUGGCUUGAAGUGAGGUUCAAUAGAGUUUGAGCCAUAGGCUUAGGCU ..((((((((((((........)))))))..((.((((.......)))).))))))).(((((.((.....)).)))))......((((((((((...)))))))))) ( -33.20, z-score = -1.15, R) >droYak2.chr2L 8622233 108 + 22324452 UUCCCUUCAUGAUUUCUCGACUCAUCAUUCAACAGAGUUAUUCACACUUGGUGUGGGUAACCAAGCUUGAAGUGAAGUUCAAUAGAGUUUGAGCCCUAGGCUUAGACU ....((((((........(((((...........))))).((((..((((((.......))))))..))))))))))........((((((((((...)))))))))) ( -31.10, z-score = -2.02, R) >droEre2.scaffold_4929 13420427 108 - 26641161 UCCCCGUCAUGAUGCCCAGAUUCAUCACAUAACAGAGGUCUUCACACUUCGUGUGGGUAACCAAGCUUGAAGUGAAGUUCAAUAGAGUUUGAGCCCUAGGCUUAUAUU .....(((.(((((........)))))........(((......(((((((..(((....)))....)))))))..((((((......))))))))).)))....... ( -25.50, z-score = -0.67, R) >droAna3.scaffold_12916 2991421 79 - 16180835 -------------------------UCCUCAUCAUGGCAC----CACUAAGUGUGGGCAACCAAGCUCGAAGUGAAGUUCAAUAGAGAUUGAGUUUCAGGCUUAGGCU -------------------------...........((.(----(((.....))))))..(((((((.....((((..(((((....)))))..))))))))).)).. ( -21.60, z-score = -0.83, R) >droVir3.scaffold_12963 17251901 80 + 20206255 -------------------------UGGGGUACAUUUAUUGCGAAAUUUAGUUCAGUUAAU-GAGCAUGCAUAGAAACUCGUUCUAUAAAAGCUCGUACAUUUCUU-- -------------------------....((((.........(((......))).......-((((.(..((((((.....))))))..).)))))))).......-- ( -14.30, z-score = -0.70, R) >droGri2.scaffold_15252 14683165 93 + 17193109 --------------UAUACACGAUGUGGACACCAGCAGCAACAAAAGUUUUUAUUAAUAAAGAUGCUUGAGAUGUUGUAGAUAUUAACAUUUUUCUUUGGAAGGCUU- --------------.......(((((........((((((.(((..(((((((....)))))))..)))...))))))........)))))((((....))))....- ( -12.79, z-score = 0.73, R) >consensus U_CCC_U_AUGAUGCAC_GACUCAUCAUCCAACAGAGUUCUUCACACUUAGUGUGGGUAACCAAGCUUGAAGUGAAGUUCAAUAGAGUUUGAGCCAUAGGCUUAGACU ............................................(((((.((.(((....))).))...)))))...........(((((((((.....))))))))) (-10.49 = -10.71 + 0.22)

| Location | 12,191,498 – 12,191,598 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 58.48 |
| Shannon entropy | 0.83709 |
| G+C content | 0.39180 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -10.75 |
| Energy contribution | -9.71 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 2.11 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12191498 100 + 23011544 UUCACACUUAGUGUGGGUAACCAGGCUUGAAGUGAAGUUCAAUAGAAUUUGAGCCAUAGGCUUAGACUGUUUUGUGUU---CUCCAUUUAUUUGUUUCGCUUG------------- ....((((((((.(((....))).)))..)))))((((..((((((.((((((((...)))))))).......(((..---...)))...))))))..)))).------------- ( -23.10, z-score = -0.57, R) >droSim1.chr2L 11992333 100 + 22036055 UUCACACUUAGUGUGGGUAACCAGGCUUGAAGUGAAGUUCAAUAGAGUUUGAGCCAUAGGCUUAGACUGUUUUGUGUU---CUCCAUUUAUUUGUUUCGCUUC------------- .........(((.(((....))).))).(((((((((..(((.(.((((((((((...)))))))))).).)))....---...((......)))))))))))------------- ( -28.80, z-score = -2.37, R) >droSec1.super_16 386978 100 + 1878335 UUCACACUUAGUGUGGGUAACCUGGCUUGAAGUGAGGUUCAAUAGAGUUUGAGCCAUAGGCUUAGGCUGUUUUGUGUU---CUCCAUUUAUUUGUUUCGCUUC------------- ..(((((...)))))((.(((((.((.....)).)))))(((.(.((((((((((...)))))))))).).)))....---..))..................------------- ( -26.40, z-score = -1.04, R) >droYak2.chr2L 8622273 103 + 22324452 UUCACACUUGGUGUGGGUAACCAAGCUUGAAGUGAAGUUCAAUAGAGUUUGAGCCCUAGGCUUAGACUGUAUUGUGUUUGGCUCCAUUUAUUUGUUUCGAUUC------------- ((((..((((((.......))))))..))))((.(((..(((((.((((((((((...)))))))))).)))))..))).)).....................------------- ( -32.90, z-score = -3.13, R) >droEre2.scaffold_4929 13420467 103 - 26641161 UUCACACUUCGUGUGGGUAACCAAGCUUGAAGUGAAGUUCAAUAGAGUUUGAGCCCUAGGCUUAUAUUGUUUUGUGUUCGGCUCUAUUUAUUUGUUUCGCUUC------------- (((((((...)))))))...........(((((((((...(((((((((.((((.(.((((.......)))).).)))))))))))))......)))))))))------------- ( -27.00, z-score = -2.00, R) >droAna3.scaffold_12916 2991436 98 - 16180835 ----CACUAAGUGUGGGCAACCAAGCUCGAAGUGAAGUUCAAUAGAGAUUGAGUUUCAGGCUUAGGCUGUUUUGUGUUCG-CCCCGUUCAGUUGUUCUGCCUC------------- ----(((.....)))((((....((((.(((.((((..(((((....)))))..))))(((..(.((......)).)..)-))...)))))))....))))..------------- ( -25.60, z-score = -0.49, R) >dp4.chr4_group3 5721765 110 - 11692001 -CUCCACUUAGUGCGGGUAACCAAGCU-----UGAAGUUCAAUAGAGUUUGAGUUUUAGGCUUAGGCUCUGUUUUUGUUUUUGUGUUUCGGAUGUUCUGAUGUUCUGUUCUGCCUC -........((.(((((.(((((((..-----((((....((((((((((((((.....))))))))))))))))))..)))).)))((((.....))))........))))))). ( -25.50, z-score = -0.50, R) >droPer1.super_1 2824467 110 - 10282868 -CUCCAUUUAGUGCGGGUAACCAAGCU-----UGAAGUUCAAUAGAGUUUGAGUUUUAGGCUUAGGCUCUGUUUUUGUUUUUGUGUUUCGGAUGUUCUGAUGUUCUGUUCUGCCUC -........((.(((((.(((((((..-----((((....((((((((((((((.....))))))))))))))))))..)))).)))((((.....))))........))))))). ( -25.50, z-score = -0.85, R) >droVir3.scaffold_12963 17251926 78 + 20206255 ----------GUUCAGUUAAU-GAGCAUGCAUAGAAACUCGUUCUAUAAAAGCUCGUACAUUUCUUAUUUUGUGUUCUUAAUAUUAUUU--------------------------- ----------.........((-((((.(..((((((.....))))))..).))))))................................--------------------------- ( -10.90, z-score = -0.66, R) >droGri2.scaffold_15252 14683198 83 + 17193109 ----------UUUUUAUUAAUAAAGAU--GCUUGAGAUGUUGUAGAUAUUAACAUUUUUCUUUGGAAGGCUUGAUCGGUGUUUCUAUUGGUCACC--------------------- ----------..........((((((.--....(((((((((.......))))))))))))))).......(((((((((....)))))))))..--------------------- ( -15.20, z-score = -0.71, R) >consensus _UCACACUUAGUGUGGGUAACCAAGCUUGAAGUGAAGUUCAAUAGAGUUUGAGCCUUAGGCUUAGACUGUUUUGUGUUUG_CUCCAUUUAUUUGUUUCGCUUC_____________ ..........((.(((....))).)).............(((.(.(((((((((.....))))))))).).))).......................................... (-10.75 = -9.71 + -1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:34 2011