
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,732,935 – 20,733,031 |
| Length | 96 |
| Max. P | 0.758093 |

| Location | 20,732,935 – 20,733,028 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.96 |
| Shannon entropy | 0.65847 |
| G+C content | 0.48002 |
| Mean single sequence MFE | -16.64 |
| Consensus MFE | -6.90 |
| Energy contribution | -6.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
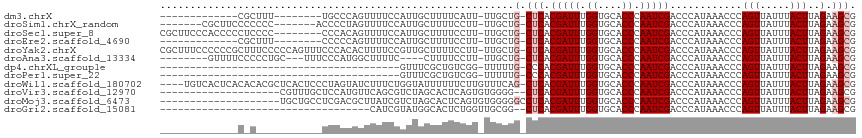
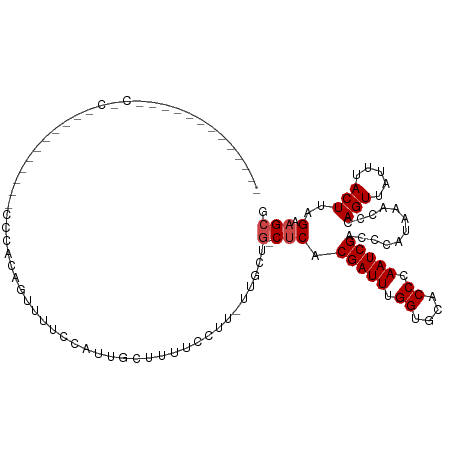
>dm3.chrX 20732935 93 - 22422827 -------------CGCUUU--------UGCCCAGUUUUCCAUUGCUUUUCAUU-UUGCUG-CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG -------------((((((--------((..((((..................-..))))-....(((((.((....)).))))).......................)))))))) ( -15.15, z-score = -1.02, R) >droSim1.chrX_random 379063 100 - 5698898 -------CGCUUCCCCCCC-------ACCCCUAGUUUUCCAUUGCUUUUCCUU-UUGCUG-CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG -------((((((......-------.................((........-..))..-....(((((.((....)).))))).........................)))))) ( -14.60, z-score = -2.16, R) >droSec1.super_8 2993860 106 - 3762037 CGCUUCCCACCCCCUCCCC--------CCCACAGUUUUCCAUUGCUUUUCCUU-UUGCUG-CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG ((((((.............--------....((((..................-..))))-....(((((.((....)).))))).........................)))))) ( -15.65, z-score = -2.46, R) >droEre2.scaffold_4690 17315911 93 - 18748788 -------------CGCUUU--------CCCCCAGUUUUCCAUUGCUUUUCCUU-UUGCUG-CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG -------------((((((--------....((((..................-..))))-....(((((.((....)).))))).........................)))))) ( -13.25, z-score = -1.13, R) >droYak2.chrX 19949587 114 - 21770863 CGCUUUCCCCCCGCUUUCCCCCAGUUUCCCACACUUUUCCGUUGCUUUUCCUU-UUGCUG-CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG ...........((((((.....(((.....((........((.((........-..)).)-)...(((((.((....)).))))).............)).....)))..)))))) ( -14.60, z-score = -1.17, R) >droAna3.scaffold_13334 53707 99 + 1562580 --------GUUUUCCCCCUGC---UUUCCCAUGGCUUUUC----CUUUUCCUU-UUGCUG-CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG --------...........((---(((.....(((.....----.........-..))).-....(((((.((....)).))))).........................))))). ( -12.49, z-score = -0.37, R) >dp4.chrXL_group1e 9377356 74 - 12523060 ----------------------------------------GUUUCGCUGUCGG-UUUUUG-CCCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG ----------------------------------------(((((...(((((-((..((-((((.....))).)))...)))))))..........(((.....)))..))))). ( -17.70, z-score = -2.47, R) >droPer1.super_22 859724 74 + 1688296 ----------------------------------------GUUUCGCUGUCGG-UUUUUG-CCCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG ----------------------------------------(((((...(((((-((..((-((((.....))).)))...)))))))..........(((.....)))..))))). ( -17.70, z-score = -2.47, R) >droWil1.scaffold_180702 1961697 111 + 4511350 ----UGUCACUCACACACGCUCACUCCCUAGUAUCUUUCUGGUAUUUUUUCUUGUUUCAG-CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG ----.............((((...((...((((..(..((((...........((....)-)...(((((.((....)).)))))..........))))..)..))))..)))))) ( -17.60, z-score = -1.15, R) >droVir3.scaffold_12970 5051617 94 - 11907090 --------------------CGUUUGCUCCAUGUUCAGCGUCUAGCACUCAGUGUGGGG--CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG --------------------(((((((((((..((.((.(.....).)).))..)))))--)...(((((.((....)).)))))..........................))))) ( -23.00, z-score = -0.69, R) >droMoj3.scaffold_6473 4567772 96 - 16943266 --------------------UGCUGCCUCGACGCUUAUCGUCUAGCACUCAGUGUGGGGGCCUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG --------------------.((((((((.(((((....(.....)....))))).)))))....(((((.((....)).)))))...........................))). ( -23.90, z-score = -0.26, R) >droGri2.scaffold_15081 3987386 79 + 4274704 -----------------------------------CAUCGUAUGGCACUCUGGUUGCGG--CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG -----------------------------------.....(((((...((.(((((.((--..(((......)))..))))))))).)))))........................ ( -14.10, z-score = 0.73, R) >consensus _____________C_C___________CCCACAGUUUUCCAUUGCUUUUCCUU_UUGCUG_CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUAGAAGCG .................................................................(((((.((....)).)))))............................... ( -6.90 = -6.90 + -0.00)

| Location | 20,732,941 – 20,733,031 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 65.06 |
| Shannon entropy | 0.70043 |
| G+C content | 0.46651 |
| Mean single sequence MFE | -13.87 |
| Consensus MFE | -6.90 |
| Energy contribution | -6.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
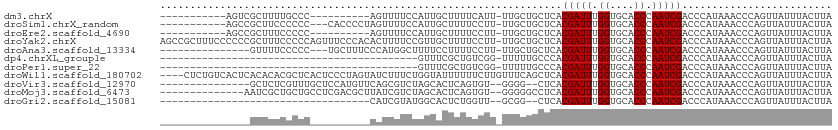
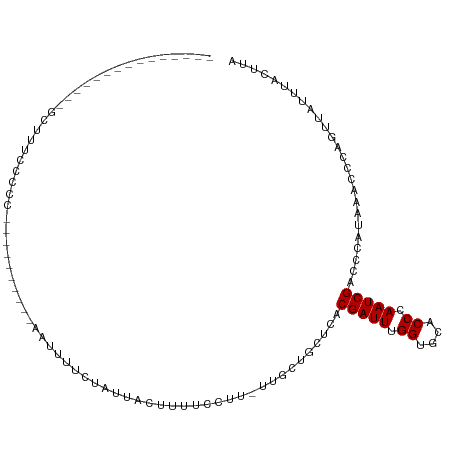
>dm3.chrX 20732941 90 - 22422827 -----------AGUCGCUUUUGCCC----------AGUUUUCCAUUGCUUUUCAUU-UUGCUGCUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA -----------.((((....(((((----------((...((....((........-.....))....)).)))).)))......))))....................... ( -11.52, z-score = -0.32, R) >droSim1.chrX_random 379069 97 - 5698898 -----------AGCCGCUUCCCCCC---CACCCCUAGUUUUCCAUUGCUUUUCCUU-UUGCUGCUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA -----------(((.((........---............................-..)).)))..(((((.((....)).)))))......................... ( -9.06, z-score = -0.36, R) >droEre2.scaffold_4690 17315917 90 - 18748788 -----------AGCCGCUUUCCCCC----------AGUUUUCCAUUGCUUUUCCUU-UUGCUGCUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA -----------(((.((........----------(((........))).......-..)).)))..(((((.((....)).)))))......................... ( -11.23, z-score = -1.00, R) >droYak2.chrX 19949593 111 - 21770863 AGCCGCUUUCCCCCCGCUUUCCCCCAGUUUCCCACACUUUUCCGUUGCUUUUCCUU-UUGCUGCUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA (((.((.........((........(((.......)))........))........-..)).)))..(((((.((....)).)))))......................... ( -10.90, z-score = -0.44, R) >droAna3.scaffold_13334 53713 93 + 1562580 ---------------GUUUUCCCCC---UGCUUUCCCAUGGCUUUUCCUUUUCCUU-UUGCUGCUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA ---------------.........(---((.........(((..............-..))).....(((((.((....)).)))))...........)))........... ( -8.99, z-score = 0.15, R) >dp4.chrXL_group1e 9377362 68 - 12523060 -------------------------------------------GUUUCGCUGUCGG-UUUUUGCCCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA -------------------------------------------.....((((..((-(((.((....(((((.((....)).)))))...)).))))))))).......... ( -14.30, z-score = -1.85, R) >droPer1.super_22 859730 68 + 1688296 -------------------------------------------GUUUCGCUGUCGG-UUUUUGCCCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA -------------------------------------------.....((((..((-(((.((....(((((.((....)).)))))...)).))))))))).......... ( -14.30, z-score = -1.85, R) >droWil1.scaffold_180702 1961703 108 + 4511350 ----CUCUGUCACUCACACACGCUCACUCCCUAGUAUCUUUCUGGUAUUUUUUCUUGUUUCAGCUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA ----............................((((..(..((((...........((....))...(((((.((....)).)))))..........))))..)..)))).. ( -13.10, z-score = -0.40, R) >droVir3.scaffold_12970 5051623 93 - 11907090 ---------------GCUCUCGUUUGCUCCAUGUUCAGCGUCUAGCACUCAGUGU--GGGG--CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA ---------------(((...((((((((((..((.((.(.....).)).))..)--))))--)...(((((.((....)).)))))......))))..))).......... ( -21.60, z-score = -0.71, R) >droMoj3.scaffold_6473 4567778 96 - 16943266 --------------AAUCGCUGCUGCCUCGACGCUUAUCGUCUAGCACUCAGUGU--GGGGGCCUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA --------------....((((..(((((.(((((....(.....)....)))))--.)))))....(((((.((....)).)))))...........)))).......... ( -23.50, z-score = -0.42, R) >droGri2.scaffold_15081 3987392 73 + 4274704 -----------------------------------CAUCGUAUGGCACUCUGGUU--GCGG--CUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA -----------------------------------.....(((((...((.((((--(.((--..(((......)))..))))))))).))))).................. ( -14.10, z-score = 0.15, R) >consensus _______________GCUUUCCCCC__________AAUUUUCUAUUACUUUUCCUU_UUGCUGCUCACGAUUUGGUGCACCCAAUCGACCCAUAAACCCAGUUAUUUACUUA ...................................................................(((((.((....)).)))))......................... ( -6.90 = -6.90 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:54 2011