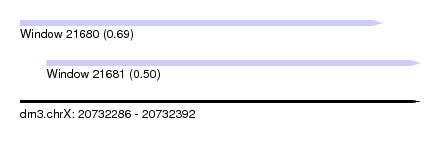
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,732,286 – 20,732,392 |
| Length | 106 |
| Max. P | 0.694251 |

| Location | 20,732,286 – 20,732,382 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.36 |
| Shannon entropy | 0.55709 |
| G+C content | 0.44361 |
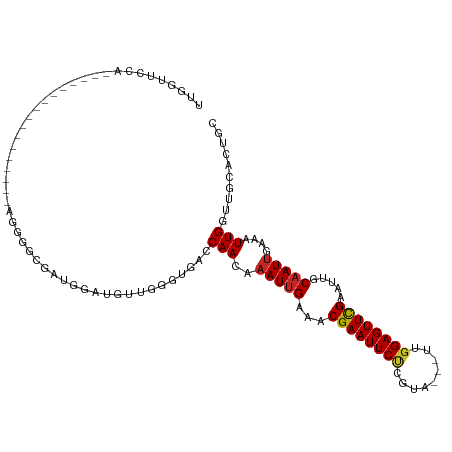
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -8.84 |
| Energy contribution | -9.05 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.694251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
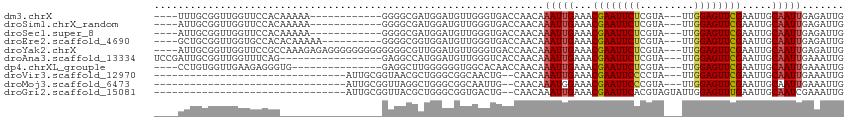
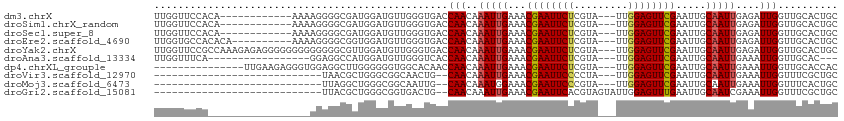
>dm3.chrX 20732286 96 + 22422827 ----UUUGCGGUUGGUUCCACAAAAA------------GGGGCGAUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUG ----.((((((((.(((((.......------------)))))......((((((....))))))........((((((((....---..))))))))))))))))......... ( -31.00, z-score = -3.10, R) >droSim1.chrX_random 378472 96 + 5698898 ----AUUGCGGUUGGUUCCACAAAAA------------GGGGCGAUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUG ----(((((((((.(((((.......------------)))))......((((((....))))))........((((((((....---..)))))))))))))))))........ ( -31.40, z-score = -3.21, R) >droSec1.super_8 2993281 96 + 3762037 ----AUUGCGGUUGGUUCCACAAAAA------------GGGGCGAUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUG ----(((((((((.(((((.......------------)))))......((((((....))))))........((((((((....---..)))))))))))))))))........ ( -31.40, z-score = -3.21, R) >droEre2.scaffold_4690 17315285 98 + 18748788 ----GCUGCGGUUGGUGCCACACAAAAA----------GGGGCGGUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUG ----..((((((((.((((...(.....----------).)))).)...((((((....))))))........((((((((....---..))))))))))))))).......... ( -29.40, z-score = -1.72, R) >droYak2.chrX 19948975 108 + 21770863 ----AUUGCGGUUGGUUCCGCCAAAGAGAGGGGGGGGGGGGGCGUUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUG ----(((((((((.(((((.((.............)).)))))......((((((....))))))........((((((((....---..)))))))))))))))))........ ( -35.42, z-score = -2.74, R) >droAna3.scaffold_13334 52411 95 - 1562580 UCCGAUUGCGGUUGGUUUCAG-----------------GAGGCCAUGGAUGUUGGGUCACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAAAUUG ..((((((((((((((((...-----------------.))))).....((((((....))))))........((((((((....---..)))))))))))))))))))...... ( -32.00, z-score = -2.88, R) >dp4.chrXL_group1e 9376853 93 + 12523060 ----CCUGUGGUUGAAGAGGGUG---------------GAGGCUUGGGGGGUGGCACAACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAAAUUG ----..((..(((...(..(((.---------------...(((........)))...)))..).........((((((((....---..)))))))))))..)).......... ( -20.00, z-score = 0.42, R) >droVir3.scaffold_12970 5051127 78 + 11907090 --------------------------------AUUGCGGUAACGCUGGGCGGCAACUG--CAACAAAUUGAAACGAAUUCCCCUA---UUGGAGUUCGAAUUGCAAUUGAAAUUG --------------------------------.(((((((..(((...)))...))))--)))..(((((...((((((((....---..)))))))).....)))))....... ( -22.70, z-score = -2.32, R) >droMoj3.scaffold_6473 4567267 78 + 16943266 --------------------------------AUUGCGGUUAGGCUGGGCGGCAAUUG--CAACAAAUGGAAACGAAUUCCCGUA---UUGGAGUUCGAAUUGCAAUUGAAAUUG --------------------------------...((.(((......))).))(((((--(((.....(....)(((((((....---..)))))))...))))))))....... ( -22.20, z-score = -1.65, R) >droGri2.scaffold_15081 3986860 81 - 4274704 --------------------------------AUUGCGGUUACGCUGGGCGGUGACUG--CAACAAAUUGAAACGAAUUCACGUAGUAUUGGAGUUUGAAUUGCAAUCGAAAUUG --------------------------------.((((((((((((...)).)))))))--)))...((((...(((((((.((......))))))))).....))))........ ( -19.60, z-score = -0.94, R) >consensus ____AUUGCGGUUGGUUCCAC_________________GGGGCGAUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA___UUGGAGUUCGAAUUGCAAUUGAAAUUG .................................................................(((((...(((((((.((......))))))))).....)))))....... ( -8.84 = -9.05 + 0.21)
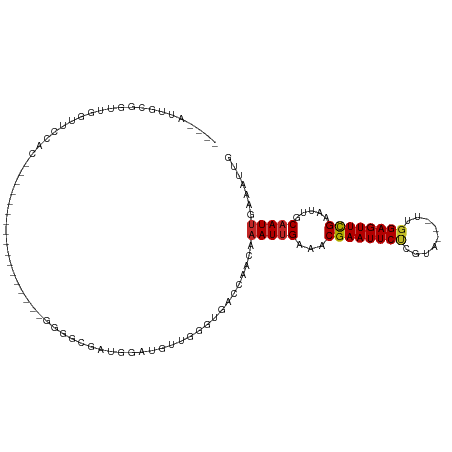
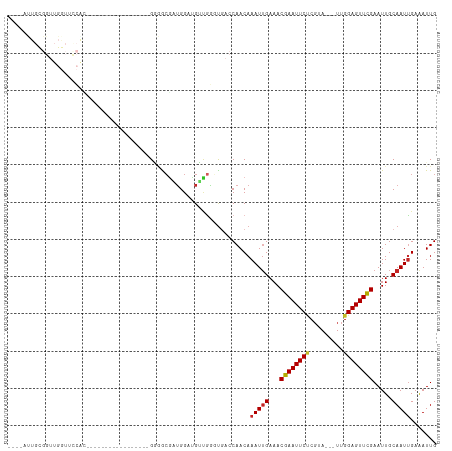
| Location | 20,732,293 – 20,732,392 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.89 |
| Shannon entropy | 0.49717 |
| G+C content | 0.44440 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -9.63 |
| Energy contribution | -9.84 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 20732293 99 + 22422827 UUGGUUCCACA------------AAAAGGGGCGAUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUGGUUGCACUGC ...(((((...------------....)))))......((((((....))))))........((((((((....---..))))))))...(((((((......))))))).... ( -30.40, z-score = -2.57, R) >droSim1.chrX_random 378479 99 + 5698898 UUGGUUCCACA------------AAAAGGGGCGAUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUGGUUGCACUGC ...(((((...------------....)))))......((((((....))))))........((((((((....---..))))))))...(((((((......))))))).... ( -30.40, z-score = -2.57, R) >droSec1.super_8 2993288 99 + 3762037 UUGGUUCCACA------------AAAAGGGGCGAUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUGGUUGCACUGC ...(((((...------------....)))))......((((((....))))))........((((((((....---..))))))))...(((((((......))))))).... ( -30.40, z-score = -2.57, R) >droEre2.scaffold_4690 17315292 101 + 18748788 UUGGUGCCACACA----------AAAAGGGGCGGUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUGGUUGCACUGC ..(((((.((.((----------(..((..(((((...((((((....))))))........((((((((....---..)))))))).)))))..))....))))).))))).. ( -33.30, z-score = -2.93, R) >droYak2.chrX 19948982 111 + 21770863 UUGGUUCCGCCAAAGAGAGGGGGGGGGGGGGCGUUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAGAUUGGUUGCACUGC ...(((((.((.............)).)))))......((((((....))))))........((((((((....---..))))))))...(((((((......))))))).... ( -34.42, z-score = -1.95, R) >droAna3.scaffold_13334 52422 91 - 1562580 UUGGUUUCA-----------------GGAGGCCAUGGAUGUUGGGUCACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAAAUUGGUUGCAC--- .((((((..-----------------..))))))....((((((....))))))........((((((((....---..))))))))...(((((((......))))))).--- ( -27.20, z-score = -2.18, R) >dp4.chrXL_group1e 9376860 96 + 12523060 ---------------UUGAAGAGGGUGGAGGCUUGGGGGGUGGCACAACCAACAAAUUGAAACGAAUUCUCGUA---UUGGAGUUCGAAUUGCAAUUGAAAUUGGUUGCACCAC ---------------........((((...(((........))).(((((((..(((((...((((((((....---..)))))))).....)))))....))))))))))).. ( -27.30, z-score = -1.90, R) >droVir3.scaffold_12970 5051134 81 + 11907090 ----------------------------UAACGCUGGGCGGCAACUG--CAACAAAUUGAAACGAAUUCCCCUA---UUGGAGUUCGAAUUGCAAUUGAAAUUGGUUUCGCUGC ----------------------------.........(((((...((--((((.....)...((((((((....---..))))))))..)))))...((((....))))))))) ( -21.40, z-score = -1.47, R) >droMoj3.scaffold_6473 4567274 81 + 16943266 ----------------------------UUAGGCUGGGCGGCAAUUG--CAACAAAUGGAAACGAAUUCCCGUA---UUGGAGUUCGAAUUGCAAUUGAAAUUGGUUUCACUGC ----------------------------..((((..(....((((((--(((.....(....)(((((((....---..)))))))...)))))))))...)..))))...... ( -24.80, z-score = -2.47, R) >droGri2.scaffold_15081 3986867 84 - 4274704 ----------------------------UUACGCUGGGCGGUGACUG--CAACAAAUUGAAACGAAUUCACGUAGUAUUGGAGUUUGAAUUGCAAUCGAAAUUGGUUUCGCUGC ----------------------------.........(((((((.((--((((.....)...(((((((.((......)))))))))..))))).))((((....))))))))) ( -17.10, z-score = 0.45, R) >consensus UUGGUUCCA_________________AGGGGCGAUGGAUGUUGGGUGACCAACAAAUUGAAACGAAUUCUCGUA___UUGGAGUUCGAAUUGCAAUUGAAAUUGGUUGCACUGC .................................................(((..(((((...(((((((.((......))))))))).....)))))....))).......... ( -9.63 = -9.84 + 0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:53 2011