| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,685,203 – 20,685,300 |
| Length | 97 |
| Max. P | 0.819734 |

| Location | 20,685,203 – 20,685,300 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.63 |
| Shannon entropy | 0.45204 |
| G+C content | 0.48625 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -13.33 |
| Energy contribution | -13.36 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
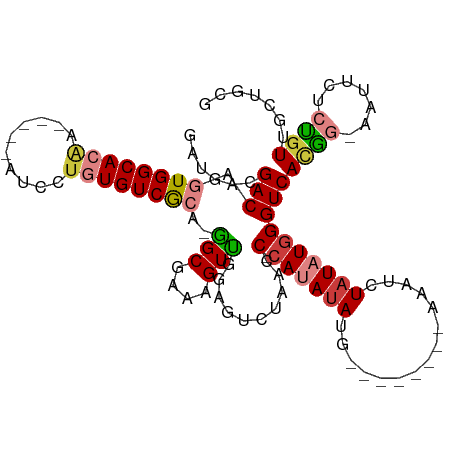
>dm3.chrX 20685203 97 + 22422827 GAUGACGACAGUGGCACAA-----AUCCUGUGUCGCA-AGCGAAAGUUGGAGUCUAACC-CAUAUAUG--------AUAUCUAUAUGGGUCACGG-AAUUCUCUUUUGCUGCG ..........((((((((.-----....)))))))).-((((((((..(((.(((.(((-((((((.(--------....))))))))))...))-).))).))))))))... ( -34.50, z-score = -3.82, R) >droEre2.scaffold_4690 17275958 97 + 18748788 GAUGACGACAGUGGCACAA-----AUCCUGUGUCGCA-GGCGAAAGUUGGACUCUAACC-CAUAUAUG--------AAAUCUAUAUGGGUCACGG-AAUUCUCUGUUGCUGCG ..........((((((((.-----....)))))))).-(((((.((..(((.(((.(((-((((((.(--------....))))))))))...))-).))).)).)))))... ( -31.00, z-score = -2.51, R) >droYak2.chrX 19908863 91 + 21770863 GAUGACGACAGUGGCACAA-----AUCCGGUGUCGCA-GGCGAAAGUUGGAGUCUAACC-CAUAUAUG--------AAUUCUAUAUGGGUCACGG-AAUUCUCCGUU------ ...(((..(.(((((((..-----.....))))))).-(((....))))..)))..(((-((((((.(--------....)))))))))).((((-(....))))).------ ( -32.80, z-score = -3.24, R) >droSec1.super_8 2948394 97 + 3762037 GAUGACGACAGUGGCACAA-----AUCCUGUGUCGCA-GGCAAAAGUUGGAGUCUAACC-CAUAUAUG--------AAAUCUAUAUGGGUCAUGG-AAUGCUCUGUUGCUGCG ..........((((((((.-----....)))))))).-(((((.....(((((((((((-((((((.(--------....))))))))))..)))-...))))).)))))... ( -31.00, z-score = -2.25, R) >droSim1.chrX 15916574 97 + 17042790 GAUGACGACAGUGGCACAA-----AUCCUGUGUCGCA-GGCGAAAGUUGGAGUCUAACC-CAUAUAUG--------AAAUCUAUAUGGGUCACGG-AAUGCUCUGUUGCUGCG ...(((..(.((((((((.-----....)))))))).-(((....))))..)))..(((-((((((.(--------....)))))))))).((((-(....)))))....... ( -32.10, z-score = -2.34, R) >droPer1.super_835 7717 113 - 10716 GGCAACGACAGUGGCACGACAGUGGAAUUCUGUCAAAUGGCGGGAGUCCCCAUGUCCCCACACAUAUAGUAUGUGCACAUCUAUAUGGGUCACAGCAGUCUUGUGUUCCCGUA ((.((((((.((((...((((..(((.(((((((....))))))).)))...)))).)))).(((((((.(((....)))))))))).)))((((.....))))))).))... ( -38.80, z-score = -1.41, R) >dp4.chrXL_group1e 9335388 96 + 12523060 GGCAACGACAGUGGCACGA-------AUUCUGUCAAAUGGCGGGAGUCCCCAUGUCCCCAAACAGA----------AGCUCUAUAUGGGUCACAGCAGUCCUGUGUUCCCGUA (....)((((.(((...((-------.(((((((....))))))).)).)))))))..........----------.......((((((.(((((.....)))))..)))))) ( -28.90, z-score = -0.60, R) >consensus GAUGACGACAGUGGCACAA_____AUCCUGUGUCGCA_GGCGAAAGUUGGAGUCUAACC_CAUAUAUG________AAAUCUAUAUGGGUCACGG_AAUUCUCUGUUGCUGCG ......(((.((((((((..........))))))))..(((....)))........................................)))...................... (-13.33 = -13.36 + 0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:46 2011