
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,190,419 – 12,190,510 |
| Length | 91 |
| Max. P | 0.748984 |

| Location | 12,190,419 – 12,190,510 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.70 |
| Shannon entropy | 0.48829 |
| G+C content | 0.41868 |
| Mean single sequence MFE | -19.44 |
| Consensus MFE | -8.93 |
| Energy contribution | -8.84 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.748984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
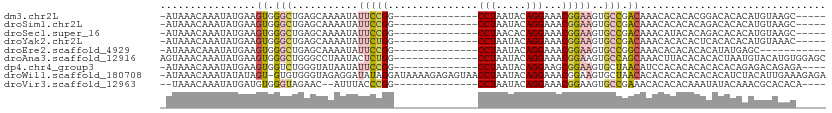
>dm3.chr2L 12190419 91 - 23011544 -AUAAACAAAUAUGAAGUGGGCUGAGCAAAAUAUUCCGG--------------CCUAAUACAGGAAACGGAAGUGCCGACAAACACACACGGACACACAUGUAAGC----- -........(((((..(((..(((.........(((((.--------------(((.....)))...)))))(((........)))...))).))).)))))....----- ( -19.10, z-score = -1.75, R) >droSim1.chr2L 11991230 91 - 22036055 -AUAAACAAAUAUGAAGUGGGCUGAGCAAAAUAUUCCGG--------------CCUAAUACAGGAAACGGAAGUGCCGACAAACACACACAGACACACAUGUAAGC----- -........(((((..(((..(((.........(((((.--------------(((.....)))...)))))(((........)))...))).))).)))))....----- ( -19.80, z-score = -2.36, R) >droSec1.super_16 385859 91 - 1878335 -AUAAACAAAUAUGAAGUGGGCUGAGCAAAAUAUUCCGG--------------CCUAACACAGGAAACGGAAGUGCCGACAAACAUACACAGACACACAUGUAAGC----- -........(((((..(((..(((.(((.....(((((.--------------(((.....)))...))))).))).(.....).....))).))).)))))....----- ( -18.30, z-score = -1.78, R) >droYak2.chr2L 8621046 91 - 22324452 -AUAAACAAAUAUGAAGUGGGCUGAGCAAAAUAUUCUGG--------------CCUAAUACAGGAAACGGAAGUGCCGACAAACACACACUCACACACAUGUAAAC----- -........(((((..(((((.((.........(((((.--------------(((.....)))...)))))(((........))).)))))))...)))))....----- ( -17.90, z-score = -1.80, R) >droEre2.scaffold_4929 13419308 85 + 26641161 -AUAAACAAAUAUGAAGUGGGCUGAGCAAAAUAUUCCGG--------------CCUAAUACAGGAAACGGAAGUGCCGGCAAACACACACACAUAUGAGC----------- -........(((((..(((.((((.(((.....(((((.--------------(((.....)))...))))).))))))).....)))...)))))....----------- ( -23.60, z-score = -3.62, R) >droAna3.scaffold_12916 2990426 97 + 16180835 AGUAAACAAAUAUGAAGUGGGCUGGGCCUAAUACUCUGG--------------CCUAAUACAGGAAACGGAAGUGCCAGCAAACUUACACACACUAAUGUACAUGUGGAGC ..............((((..((((((((.........))--------------)))......((..((....)).)))))..)))).((((.((....))...)))).... ( -22.50, z-score = -0.31, R) >dp4.chr4_group3 5720413 92 + 11692001 -AUAAACAAAUAUGAAGUGGUCUGGGUAUAAUAUUCCGG--------------CCUAAUACAGGAAGCGGAAGUGCUAACAUCCACACACACACACAGAGACAGAGA---- -...............(((((.(((((......(((((.--------------(((.....)))...)))))((....))))))).)).)))...............---- ( -18.40, z-score = -1.02, R) >droWil1.scaffold_180708 9873041 109 - 12563649 -AUAAACAAAUAUAUAGU-GUGUGGGUAGAGGAUAUAGGAUAAAAGAGAGUAACCUAAUACAGGAAACGGAAGUGCUAACACACACACACACACAUCUACAUUGAAAGAGA -...............((-(((((..(((......((((.((........)).)))).........((....)).)))...)))))))....................... ( -18.60, z-score = -1.59, R) >droVir3.scaffold_12963 17251022 89 - 20206255 --UAAACAAAUAUGAUGUGGGUAGAAC--AUUUACCCGG--------------CCUAAUACAGGAAACGGAAGUGCCGAAACACACACAAAUAUACAAACGCACACA---- --.......((((..((((((((((..--.))))))(((--------------(((......(....)...)).)))).......)))).)))).............---- ( -16.80, z-score = -1.84, R) >consensus _AUAAACAAAUAUGAAGUGGGCUGAGCAAAAUAUUCCGG______________CCUAAUACAGGAAACGGAAGUGCCGACAAACACACACACACACACAUGUAAAC_____ ................((.(((...............................(((.....)))..((....))))).))............................... ( -8.93 = -8.84 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:33 2011