
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,646,510 – 20,646,595 |
| Length | 85 |
| Max. P | 0.931722 |

| Location | 20,646,510 – 20,646,595 |
|---|---|
| Length | 85 |
| Sequences | 15 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Shannon entropy | 0.40440 |
| G+C content | 0.43861 |
| Mean single sequence MFE | -18.99 |
| Consensus MFE | -10.93 |
| Energy contribution | -11.23 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
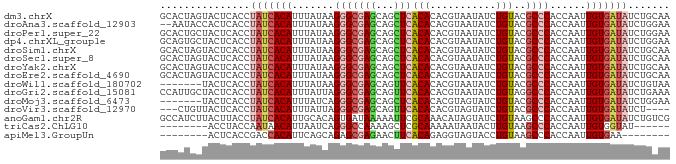
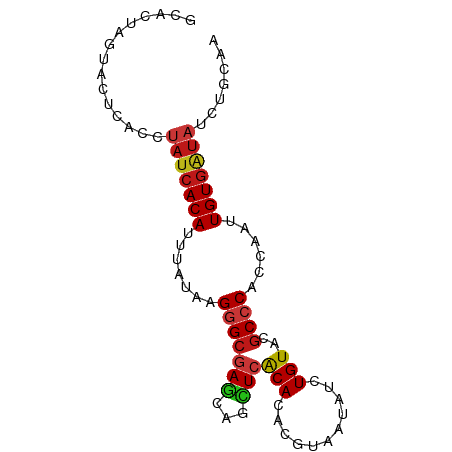
>dm3.chrX 20646510 85 + 22422827 GCACUAGUACUCACCUAUCACAUUUAUAAGGGCGAGCAGCUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGCAA (((...((....)).(((((((.......(((((.((((...((....)).....)))).)))))......)))))))..))).. ( -20.82, z-score = -2.44, R) >droAna3.scaffold_12903 759507 83 - 802071 --AAUACCACUCACCUAUCACAUUUAUAAGGGCGAGCAGCUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGGAA --........(((..(((((((.......(((((.((((...((....)).....)))).)))))......)))))))..))).. ( -21.82, z-score = -3.34, R) >droPer1.super_22 778452 85 - 1688296 GCACUGCUACUCACCUAUCACAUUUAUAAGGGCGAGCAGCUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGGAA ..........(((..(((((((.......(((((.((((...((....)).....)))).)))))......)))))))..))).. ( -21.82, z-score = -2.38, R) >dp4.chrXL_group1e 9298196 85 + 12523060 GCAGUGCUACUCACCUAUCACAUUUAUAAGGGCGAGCAGCUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGGAA ..........(((..(((((((.......(((((.((((...((....)).....)))).)))))......)))))))..))).. ( -21.82, z-score = -1.80, R) >droSim1.chrX 15875945 85 + 17042790 GCACUAGUACUCACCUAUCACAUUUAUAAGGGCGAGCAGCUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGCAA (((...((....)).(((((((.......(((((.((((...((....)).....)))).)))))......)))))))..))).. ( -20.82, z-score = -2.44, R) >droSec1.super_8 2885673 85 + 3762037 GCACUAGUACUCACCUAUCACAUUUAUAAGGGCGAGCAGCUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGCAA (((...((....)).(((((((.......(((((.((((...((....)).....)))).)))))......)))))))..))).. ( -20.82, z-score = -2.44, R) >droYak2.chrX 19871786 85 + 21770863 GCACUAGUACUCACCUAUCACAUUUAUAAGGGCGAGCAGCUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGCAA (((...((....)).(((((((.......(((((.((((...((....)).....)))).)))))......)))))))..))).. ( -20.82, z-score = -2.44, R) >droEre2.scaffold_4690 17239609 85 + 18748788 GCACUAGUACUCACCUAUCACAUUUAUAAGGGCGAGCAGCUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGCAA (((...((....)).(((((((.......(((((.((((...((....)).....)))).)))))......)))))))..))).. ( -20.82, z-score = -2.44, R) >droWil1.scaffold_180702 1886290 78 - 4511350 -------UACUCACCUAUCACAUUUAUAAGGGCGAGCAGUUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGUAA -------....((..(((((((.......(((((.((((...((....)).....)))).)))))......)))))))..))... ( -21.32, z-score = -3.64, R) >droGri2.scaffold_15081 3905802 85 - 4274704 CCAUUGCUACUCACCUAUCACAUUUAUUAGGGCGAGCAGUUCACACACGUAAUAUCUGUAGGCCCACCAAUUGUGAUAUCUGAAA ..........(((..(((((((.......((((..((((...((....)).....))))..))))......)))))))..))).. ( -20.32, z-score = -2.02, R) >droMoj3.scaffold_6473 4464182 78 + 16943266 -------UACUCACCUAUCACAUUUAUCAGGGCGAGCAGCUCACACACGUAGUAUCUGUACGCCCACCAAUUGUGAUAUCUGGAA -------...(((..(((((((.......(((((.((((..(((....)).)...)))).)))))......)))))))..))).. ( -22.52, z-score = -3.07, R) >droVir3.scaffold_12970 4972621 78 + 11907090 ---CUGUUACUCACCUAUCACAUUUAUUAGGGCGAGCAGUUCACACACGUAGUAUCUGUACGCCCACCAAUUGUGAUAUCU---- ---............(((((((.......(((((.((((..(((....)).)...)))).)))))......)))))))...---- ( -21.62, z-score = -3.31, R) >anoGam1.chr2R 5476888 85 + 62725911 GCCAUCUUACUUACCUAUCACAUUGCACAGUGAUAAAAAUUCGCAAACAUAGUAUCUGUAAGCCCACCAAUUGUGAUAUCUGUCG ...............((((((((((....((((.......))))....((((...))))........))).)))))))....... ( -10.50, z-score = -0.33, R) >triCas2.ChLG10 7098110 71 + 8806720 --------ACCUACCAAUAACAUUAAUCAGGGCCAAAAGCUCGCAAAAAUAAUACUUGUAAGCCCACCAAUUGUGGUAU------ --------.....................((((...(((...............)))....))))((((....))))..------ ( -9.56, z-score = 0.14, R) >apiMel3.GroupUn 378051989 69 - 399230636 --------ACUCACCGACCACAUUCAGCAGAGCGAGAACUUCACAGAGGUAGUACCUGUAAGCCCACCAAUUGUGAA-------- --------.(((...((......)).((...)))))....((((((.(((.((........))..)))..)))))).-------- ( -9.40, z-score = 0.66, R) >consensus GCACUAGUACUCACCUAUCACAUUUAUAAGGGCGAGCAGCUCACACACGUAAUAUCUGUACGCCCACCAAUUGUGAUAUCUGCAA ...............(((((((.......(((((((...)))(((...........)))..))))......)))))))....... (-10.93 = -11.23 + 0.30)
| Location | 20,646,510 – 20,646,595 |
|---|---|
| Length | 85 |
| Sequences | 15 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 82.94 |
| Shannon entropy | 0.40440 |
| G+C content | 0.43861 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -15.47 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
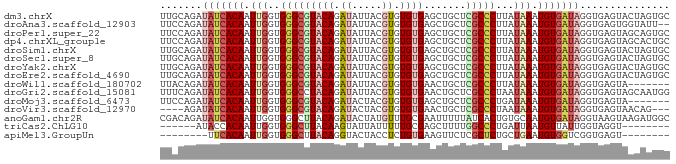
>dm3.chrX 20646510 85 - 22422827 UUGCAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAGCUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUACUAGUGC (..(...(((((((.(((..(((((..(((...((((....)))).)))..)))))...))).))))))).)..).......... ( -28.40, z-score = -2.30, R) >droAna3.scaffold_12903 759507 83 + 802071 UUCCAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAGCUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUGGUAUU-- ...((..(((((((.(((..(((((..(((...((((....)))).)))..)))))...))).)))))))..)).........-- ( -28.10, z-score = -2.76, R) >droPer1.super_22 778452 85 + 1688296 UUCCAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAGCUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUAGCAGUGC ...((..(((((((.(((..(((((..(((...((((....)))).)))..)))))...))).)))))))..))........... ( -28.10, z-score = -2.23, R) >dp4.chrXL_group1e 9298196 85 - 12523060 UUCCAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAGCUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUAGCACUGC .......(((((((.(((..(((((..(((...((((....)))).)))..)))))...))).)))))))((((.....)))).. ( -28.60, z-score = -2.29, R) >droSim1.chrX 15875945 85 - 17042790 UUGCAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAGCUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUACUAGUGC (..(...(((((((.(((..(((((..(((...((((....)))).)))..)))))...))).))))))).)..).......... ( -28.40, z-score = -2.30, R) >droSec1.super_8 2885673 85 - 3762037 UUGCAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAGCUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUACUAGUGC (..(...(((((((.(((..(((((..(((...((((....)))).)))..)))))...))).))))))).)..).......... ( -28.40, z-score = -2.30, R) >droYak2.chrX 19871786 85 - 21770863 UUGCAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAGCUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUACUAGUGC (..(...(((((((.(((..(((((..(((...((((....)))).)))..)))))...))).))))))).)..).......... ( -28.40, z-score = -2.30, R) >droEre2.scaffold_4690 17239609 85 - 18748788 UUGCAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAGCUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUACUAGUGC (..(...(((((((.(((..(((((..(((...((((....)))).)))..)))))...))).))))))).)..).......... ( -28.40, z-score = -2.30, R) >droWil1.scaffold_180702 1886290 78 + 4511350 UUACAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAACUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUA------- ((((...(((((((.(((..(((((..(((...((((....)))).)))..)))))...))).))))))).))))...------- ( -26.80, z-score = -3.32, R) >droGri2.scaffold_15081 3905802 85 + 4274704 UUUCAGAUAUCACAAUUGGUGGGCCUACAGAUAUUACGUGUGUGAACUGCUCGCCCUAAUAAAUGUGAUAGGUGAGUAGCAAUGG .((((..(((((((.(((..((((...(((...((((....)))).)))...))))...))).)))))))..))))......... ( -23.60, z-score = -1.50, R) >droMoj3.scaffold_6473 4464182 78 - 16943266 UUCCAGAUAUCACAAUUGGUGGGCGUACAGAUACUACGUGUGUGAGCUGCUCGCCCUGAUAAAUGUGAUAGGUGAGUA------- ...((..(((((((.(((..(((((..(((((((.....))))...)))..)))))...))).)))))))..))....------- ( -26.60, z-score = -2.43, R) >droVir3.scaffold_12970 4972621 78 - 11907090 ----AGAUAUCACAAUUGGUGGGCGUACAGAUACUACGUGUGUGAACUGCUCGCCCUAAUAAAUGUGAUAGGUGAGUAACAG--- ----...(((((((.(((..(((((..(((((((.....))))...)))..)))))...))).)))))))............--- ( -24.50, z-score = -2.58, R) >anoGam1.chr2R 5476888 85 - 62725911 CGACAGAUAUCACAAUUGGUGGGCUUACAGAUACUAUGUUUGCGAAUUUUUAUCACUGUGCAAUGUGAUAGGUAAGUAAGAUGGC .......((((..((((.(..(((.............)))..).))))((((((((........)))))))).......)))).. ( -15.22, z-score = 0.50, R) >triCas2.ChLG10 7098110 71 - 8806720 ------AUACCACAAUUGGUGGGCUUACAAGUAUUAUUUUUGCGAGCUUUUGGCCCUGAUUAAUGUUAUUGGUAGGU-------- ------.(((((..(((((((((((...((((.............))))..)))))..)))))).....)))))...-------- ( -15.42, z-score = -0.20, R) >apiMel3.GroupUn 378051989 69 + 399230636 --------UUCACAAUUGGUGGGCUUACAGGUACUACCUCUGUGAAGUUCUCGCUCUGCUGAAUGUGGUCGGUGAGU-------- --------((((((...((((((((....))).)))))..))))))...(((((((..(.....)..)..)))))).-------- ( -22.30, z-score = -1.83, R) >consensus UUGCAGAUAUCACAAUUGGUGGGCGUACAGAUAUUACGUGUGUGAGCUGCUCGCCCUUAUAAAUGUGAUAGGUGAGUACUAGUGC .......(((((((.(((..(((((((((.((.....)).)))).......)))))...))).)))))))............... (-15.47 = -16.05 + 0.58)
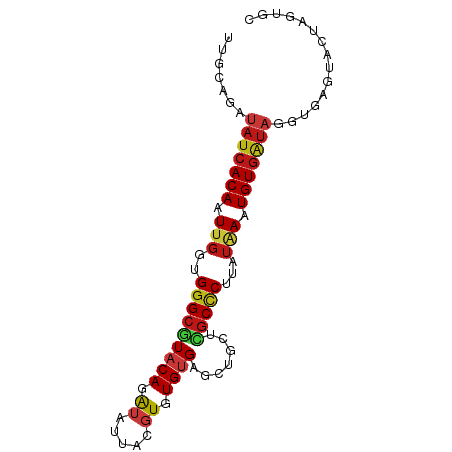
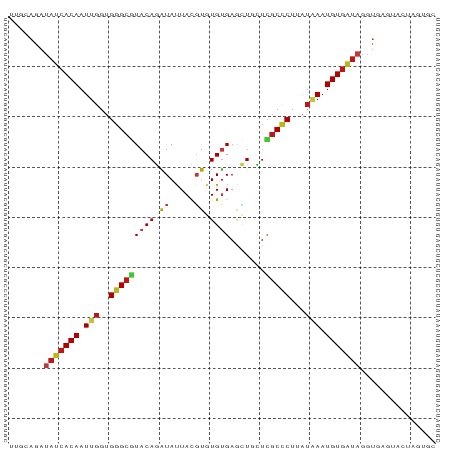
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:43 2011