| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,645,830 – 20,645,924 |
| Length | 94 |
| Max. P | 0.603478 |

| Location | 20,645,830 – 20,645,924 |
|---|---|
| Length | 94 |
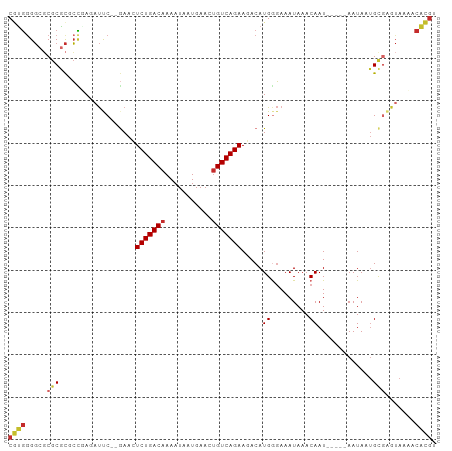
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Shannon entropy | 0.35390 |
| G+C content | 0.41348 |
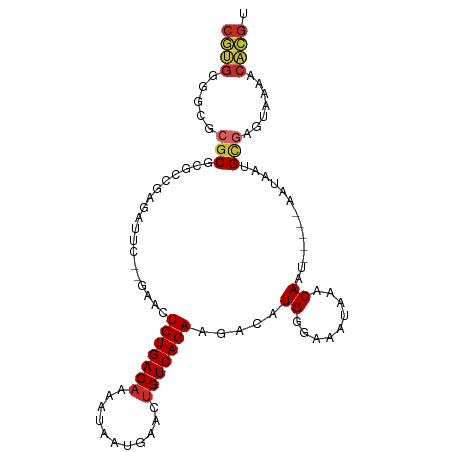
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -10.87 |
| Energy contribution | -11.08 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20645830 94 - 22422827 CGUGGGGCGCGCGCGCCUAGAUUC--GAACUCUGACAAAAUAAUGAACUGUCAGAAGACAUGGGAAAUAAACAAU-----AAUAUUGCGAGUAAAACACGU (((((((((....)))))..((((--(...(((((((...........)))))))................((((-----...)))))))))....)))). ( -23.60, z-score = -2.05, R) >droSim1.chrX 15875257 94 - 17042790 CGUGAGGCGCGCGCGCCUAGAUUC--GAACUCUGACAAAAUAAUGAACUGUCAGAAGACAUGGGAAAUAAACAAU-----AAUAAUGCGAGUAAAACACGU (((((((((....)))))..((((--(...(((((((...........))))))).....((.........))..-----.......)))))....)))). ( -23.30, z-score = -2.38, R) >droSec1.super_8 2884985 94 - 3762037 CGUGGGGCGCGCGCGCCUAGAUUC--GAACUCUGACAAAAUAAUGAACUGUCAGAAGACAUGGGAAAUAAACAAU-----AAUAAUGCGAGUAAAACACGU (((((((((....)))))..((((--(...(((((((...........))))))).....((.........))..-----.......)))))....)))). ( -23.50, z-score = -2.20, R) >droYak2.chrX 19871051 94 - 21770863 CGUGGGGCGCGCGCGCCGAGCUUC--GAACUCUGACAAAAUAAUGAACUGUCAGAAGACAUGGGAAAUAAACAAU-----AAUAUUGUGAGUAAAACACGU ((((.((((....))))..((((.--....(((((((...........)))))))...............(((((-----...)))))))))....)))). ( -21.90, z-score = -0.98, R) >droEre2.scaffold_4690 17238863 99 - 18748788 CGUGGGGCGCGCGCGCCGAGAUUC--GAACUCUGACAAAAUAAUGAACUGUCAGAAGACAUGGGAAAUAAACAAUAAUACAAUAUUGCGAGUAAAACACGU ((((.((((....))))...((((--(...(((((((...........)))))))................(((((......))))))))))....)))). ( -25.00, z-score = -2.29, R) >droWil1.scaffold_180702 1885818 84 + 4511350 CAUUGGCCCCACAUUGAAAUCCCCAAAAAUUCUGACAAAAUAAUGAACAGUCAGAAAACAUGAUAAAUAAACAAU-----AAUA--GCCAA---------- ..(((((......(((.......)))...(((((((.............)))))))...................-----....--)))))---------- ( -12.52, z-score = -2.96, R) >droAna3.scaffold_12903 759064 92 + 802071 CGUGGGG--CGCACGCUGAAAUCC--GAAAUCUGACAAAAUAAUGAACUGUCAGAAAACAUGAGAAAUAAACAAU-----AAUAAUGCGAGUAAUACACGU ((((...--.((.(((........--....(((((((...........))))))).......(....).......-----......))).))....)))). ( -15.80, z-score = -1.08, R) >droPer1.super_22 777608 94 + 1688296 CUCGGGGCGCGCGCUCUGAAAUCC--GAAAUCUGACAAAAUAAUGAACUGUCAGAAAACAUGGGAAAUAAACAAU-----AAUAAUGCCGAUAAUGCGGGU .(((((((....))))))).((((--(...(((((((...........)))))))....((.((...((......-----..))...)).))....))))) ( -22.90, z-score = -2.11, R) >dp4.chrXL_group1e 9297379 94 - 12523060 CUCGGGGCGCGCGCUCUGAAAUCC--AAAAUCUGACAAAAUAAUGAACUGUCAGAAAACAUGGGAAAUAAACAAU-----AAUAAUGCGGAUAAUGCGGGU .(((((((....))))))).((((--....(((((((...........)))))))....................-----......(((.....))))))) ( -22.70, z-score = -2.54, R) >consensus CGUGGGGCGCGCGCGCCGAGAUUC__GAACUCUGACAAAAUAAUGAACUGUCAGAAGACAUGGGAAAUAAACAAU_____AAUAAUGCGAGUAAAACACGU ((((.....(((..................(((((((...........))))))).....((.........)).............))).......)))). (-10.87 = -11.08 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:40 2011