| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,643,027 – 20,643,117 |
| Length | 90 |
| Max. P | 0.996393 |

| Location | 20,643,027 – 20,643,117 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 55.60 |
| Shannon entropy | 0.86774 |
| G+C content | 0.59946 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -8.23 |
| Energy contribution | -8.62 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.77 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
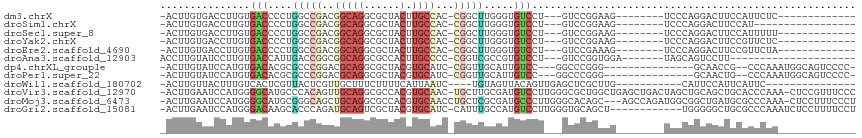
>dm3.chrX 20643027 90 + 22422827 -ACUUGUGACCUUGUGACCCCUGGCCGACGGCAGGCGCUACUUGCCAC-CGGCUUGGGUGUCCU---GUCCGGAAG--------UCCCAGGACUUCCAUUCUC------------- -....(.(((...(.((((((.(((((..((((((.....))))))..-))))).))).)))).---))))(((((--------((....)))))))......------------- ( -42.60, z-score = -3.95, R) >droSim1.chrX 15872017 86 + 17042790 -ACUUGUGACCUUGUGACCCCUGGCCGACGGCAGGCGCUACUUGCCAC-CGGCUUGGGUGUCCU---GUCCGGAAG--------UCCCAGGACUUCCAU----------------- -....(.(((...(.((((((.(((((..((((((.....))))))..-))))).))).)))).---))))(((((--------((....)))))))..----------------- ( -42.60, z-score = -4.04, R) >droSec1.super_8 2882153 90 + 3762037 -ACUUGUGACCUUGUGACCCCUGGCCGACGGCAGGCGCUACUUGCCAC-CGGCUUGGGUGUCCU---GUCCGGAAG--------UCCCAGGACUUCCAUUUUU------------- -....(.(((...(.((((((.(((((..((((((.....))))))..-))))).))).)))).---))))(((((--------((....)))))))......------------- ( -42.60, z-score = -4.26, R) >droYak2.chrX 19868143 90 + 21770863 -ACUUGUGACCUUGUGACCCCUGGCCGACGGCAGGCGCUACUUGCCAC-CGGCUUGGGUGUCCU---GUCCGGAAG--------UCCCAGGACUUCCGUUCUC------------- -......(((...(.((((((.(((((..((((((.....))))))..-))))).))).)))).---)))((((((--------((....)))))))).....------------- ( -43.50, z-score = -3.90, R) >droEre2.scaffold_4690 17235997 90 + 18748788 -ACUUGUGACCUUGUGACCCCUGGCCGACGGCAGGCGCUACUUGCCAC-CGGCUUGGGUGUCCU---GUCCGAAAG--------UCCCAGGACUUCCGUUCUA------------- -......(((...(.((((((.(((((..((((((.....))))))..-))))).))).)))).---)))((.(((--------((....))))).)).....------------- ( -36.70, z-score = -2.50, R) >droAna3.scaffold_12903 756259 84 - 802071 ACCUUGUAUCCUUGUGACCAUUGACCGGCGGCAGGCGCCACUUGCCCC-CGGUCGCCGUGUCCU---GUCCGGUGGA-------UAGCAGUCCUU--------------------- ...((((((((...........((((((.((((((.....)))))).)-)))))((((.(....---..))))))))-------).)))).....--------------------- ( -32.10, z-score = -1.64, R) >dp4.chrXL_group1e 9295020 93 + 12523060 -ACUUGUAUCCAUGUGACACGCGCCCGGACGCAGGCGCUACGUGCAUC-CGGUUGCAUUGUCC---GGCCCGGG---------------GCAACCG--CCCAAAUGGCAGUCCCC- -...........((.((((.(((.(((((.(((.(.....).))).))-))).)))..)))))---)(((..((---------------((....)--)))....))).......- ( -33.60, z-score = 0.52, R) >droPer1.super_22 775235 93 - 1688296 -ACUUGUAUCCAUGUGACACGCGCCCGGACGCAGGCGCUACGUGCAUC-CGGUUGCAUUGUCC---GGCCCGGG---------------GCAACUG--CCCAAAUGGCAGUCCCC- -...........((.((((.(((.(((((.(((.(.....).))).))-))).)))..)))))---)....(((---------------(..((((--((.....))))))))))- ( -36.40, z-score = -0.41, R) >droWil1.scaffold_180702 1883116 85 - 4511350 -ACUUGUUACUUUGUCACUCGUUACUCGUUGCUUUCUUUUCAUUAAUC----UGUAGUUACAGUUGAGCUCGCUUG-----------CAUUCCAUUCAUUC--------------- -...................((....((..((((.............(----(((....))))..)))).))...)-----------).............--------------- ( -5.86, z-score = 1.31, R) >droVir3.scaffold_12970 4969831 113 + 11907090 -ACUUGAAUCCAUGGGGCAUGCCCACAGUUGCAGGCGCCACGUGCAAC-UGCUUGCGAUGUCCUUGGGCGCUGGCUGAGCUGACUAGCUGCAGCUGCACCCAAA-CUCCGUUUCCC -.............((((((((...((((((((.(.....).))))))-))...)).))))))(((((.((.(((((((((....)))).))))))).))))).-........... ( -44.70, z-score = -1.28, R) >droMoj3.scaffold_6473 4460835 111 + 16943266 -ACUUGAAUCCAUGGGGCAUGCGGGCAGCUGCAGGCGCCACGUGCAACCUGCUCGCGAUGCCCUUGGGCACAGC---AGCCAGAUGGCGGCUGAUGCGCCCAAA-CUCCUUUCCCU -.............((((((((((((((.((((.(.....).))))..)))))))).))))))((((((.((.(---((((.......))))).)).)))))).-........... ( -52.10, z-score = -1.89, R) >droGri2.scaffold_15081 3903039 102 - 4274704 -ACUUGAAUCCAUGGGACAAGCACCCAGAUGCAGUCGCUACGUGCAUC-CAUUUGCCAUGUCCUUGGGUGCAGCU------------UGGGGGCUGCGCCCAAAUCUCCUUUUCCU -.............(((((.(((....((((((((....)).))))))-....)))..)))))((((((((((((------------....))))))))))))............. ( -38.80, z-score = -2.11, R) >consensus _ACUUGUAACCUUGUGACCCGUGGCCGGCGGCAGGCGCUACUUGCAAC_CGGUUGCGGUGUCCU___GUCCGGAAG________UCCCAGGACUUCCAUCCAA_____________ ...............(((....(((((..(((((......).))))...))))).....)))...................................................... ( -8.23 = -8.62 + 0.39)

| Location | 20,643,027 – 20,643,117 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 55.60 |
| Shannon entropy | 0.86774 |
| G+C content | 0.59946 |
| Mean single sequence MFE | -40.98 |
| Consensus MFE | -13.01 |
| Energy contribution | -11.72 |
| Covariance contribution | -1.29 |
| Combinations/Pair | 2.12 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
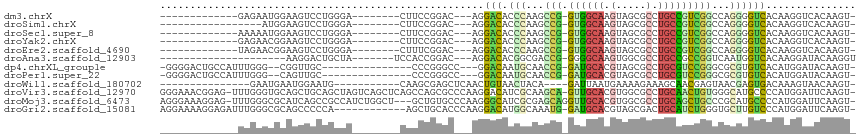
>dm3.chrX 20643027 90 - 22422827 -------------GAGAAUGGAAGUCCUGGGA--------CUUCCGGAC---AGGACACCCAAGCCG-GUGGCAAGUAGCGCCUGCCGUCGGCCAGGGGUCACAAGGUCACAAGU- -------------.....((((((((....))--------))))))(((---..(((.(((..((((-..((((.(.....).))))..))))..)))))).....)))......- ( -42.70, z-score = -3.29, R) >droSim1.chrX 15872017 86 - 17042790 -----------------AUGGAAGUCCUGGGA--------CUUCCGGAC---AGGACACCCAAGCCG-GUGGCAAGUAGCGCCUGCCGUCGGCCAGGGGUCACAAGGUCACAAGU- -----------------.((((((((....))--------))))))(((---..(((.(((..((((-..((((.(.....).))))..))))..)))))).....)))......- ( -42.70, z-score = -3.50, R) >droSec1.super_8 2882153 90 - 3762037 -------------AAAAAUGGAAGUCCUGGGA--------CUUCCGGAC---AGGACACCCAAGCCG-GUGGCAAGUAGCGCCUGCCGUCGGCCAGGGGUCACAAGGUCACAAGU- -------------.....((((((((....))--------))))))(((---..(((.(((..((((-..((((.(.....).))))..))))..)))))).....)))......- ( -42.70, z-score = -3.72, R) >droYak2.chrX 19868143 90 - 21770863 -------------GAGAACGGAAGUCCUGGGA--------CUUCCGGAC---AGGACACCCAAGCCG-GUGGCAAGUAGCGCCUGCCGUCGGCCAGGGGUCACAAGGUCACAAGU- -------------.....((((((((....))--------))))))(((---..(((.(((..((((-..((((.(.....).))))..))))..)))))).....)))......- ( -44.90, z-score = -3.86, R) >droEre2.scaffold_4690 17235997 90 - 18748788 -------------UAGAACGGAAGUCCUGGGA--------CUUUCGGAC---AGGACACCCAAGCCG-GUGGCAAGUAGCGCCUGCCGUCGGCCAGGGGUCACAAGGUCACAAGU- -------------.....((((((((....))--------))))))(((---..(((.(((..((((-..((((.(.....).))))..))))..)))))).....)))......- ( -42.00, z-score = -3.58, R) >droAna3.scaffold_12903 756259 84 + 802071 ---------------------AAGGACUGCUA-------UCCACCGGAC---AGGACACGGCGACCG-GGGGCAAGUGGCGCCUGCCGCCGGUCAAUGGUCACAAGGAUACAAGGU ---------------------....(((..((-------(((....(((---..(((.(((((..((-((.((.....)).)))).))))))))....)))....)))))...))) ( -33.40, z-score = -1.43, R) >dp4.chrXL_group1e 9295020 93 - 12523060 -GGGGACUGCCAUUUGGG--CGGUUGC---------------CCCGGGCC---GGACAAUGCAACCG-GAUGCACGUAGCGCCUGCGUCCGGGCGCGUGUCACAUGGAUACAAGU- -((((((((((.....))--))))..)---------------)))(..((---.((((.(((..(((-((((((.(.....).)))))))))..)))))))....))...)....- ( -43.70, z-score = -1.31, R) >droPer1.super_22 775235 93 + 1688296 -GGGGACUGCCAUUUGGG--CAGUUGC---------------CCCGGGCC---GGACAAUGCAACCG-GAUGCACGUAGCGCCUGCGUCCGGGCGCGUGUCACAUGGAUACAAGU- -((((((((((.....))--))))..)---------------)))(..((---.((((.(((..(((-((((((.(.....).)))))))))..)))))))....))...)....- ( -44.40, z-score = -1.86, R) >droWil1.scaffold_180702 1883116 85 + 4511350 ---------------GAAUGAAUGGAAUG-----------CAAGCGAGCUCAACUGUAACUACA----GAUUAAUGAAAAGAAAGCAACGAGUAACGAGUGACAAAGUAACAAGU- ---------------............((-----------((..((.((((..((((....)))----).....(....).........))))..))..)).))...........- ( -9.30, z-score = 0.65, R) >droVir3.scaffold_12970 4969831 113 - 11907090 GGGAAACGGAG-UUUGGGUGCAGCUGCAGCUAGUCAGCUCAGCCAGCGCCCAAGGACAUCGCAAGCA-GUUGCACGUGGCGCCUGCAACUGUGGGCAUGCCCCAUGGAUUCAAGU- .(....)((..-.((((((((.((((.((((....))))))))..))))))))((.(((.((..(((-((((((.(.....).)))))))))..)))))))))............- ( -49.10, z-score = -1.53, R) >droMoj3.scaffold_6473 4460835 111 - 16943266 AGGGAAAGGAG-UUUGGGCGCAUCAGCCGCCAUCUGGCU---GCUGUGCCCAAGGGCAUCGCGAGCAGGUUGCACGUGGCGCCUGCAGCUGCCCGCAUGCCCCAUGGAUUCAAGU- ........(((-((((((((((.((((((.....)))))---).)))))))..((((((.(((.((((.(((((.(.....).))))))))).)))))))))...))))))....- ( -55.20, z-score = -1.76, R) >droGri2.scaffold_15081 3903039 102 + 4274704 AGGAAAAGGAGAUUUGGGCGCAGCCCCCA------------AGCUGCACCCAAGGACAUGGCAAAUG-GAUGCACGUAGCGACUGCAUCUGGGUGCUUGUCCCAUGGAUUCAAGU- ..(((........(((((.(((((.....------------.))))).)))))(((((.((((..((-((((((.((....))))))))))..)))))))))......)))....- ( -41.60, z-score = -2.29, R) >consensus _____________UUGAAUGCAAGUCCUGGGA________CUUCCGGAC___AGGACACCGAAACCG_GUGGCAAGUAGCGCCUGCCGCCGGCCACGGGUCACAAGGAUACAAGU_ ......................................................(((.(((...(((.((((((.(.....).)))))))))...))))))............... (-13.01 = -11.72 + -1.29)
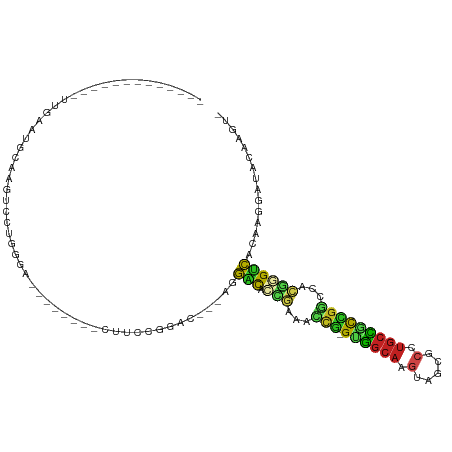
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:39 2011