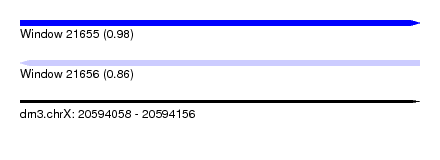
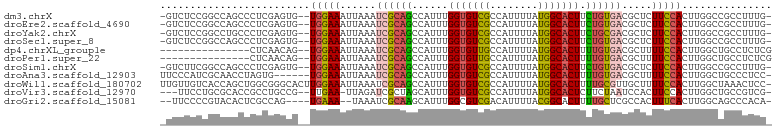
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,594,058 – 20,594,156 |
| Length | 98 |
| Max. P | 0.978553 |

| Location | 20,594,058 – 20,594,156 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.96 |
| Shannon entropy | 0.52764 |
| G+C content | 0.51026 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.69 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
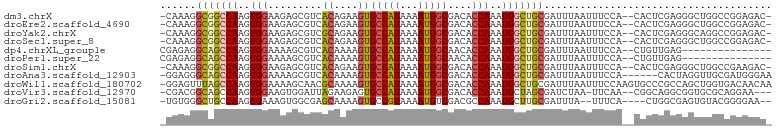
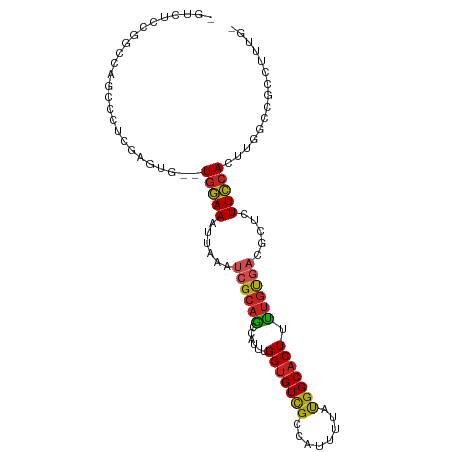
>dm3.chrX 20594058 98 + 22422827 -CAAAGGCGGCCAAGUGGAAGAGCGUCACAGAAGUGCCAUAAAAUGGCGACACCAAAUGGCUGCGAUUUAAUUUCCA--CACUCGAGGGCUGGCCGGAGAC- -......((((((.(((((((...(((.(((..(((((((...))))).)).((....))))).)))....))))))--).((....)).)))))).....- ( -31.80, z-score = -1.68, R) >droEre2.scaffold_4690 17189425 98 + 18748788 -CAAAGGCGGCCAAGUGGAAGAGCGUCACAGAAGUGCCAUAAAAUGGCGACACCAAAUGGCUGCGAUUUAAUUUCCA--CACUCGAGGGCUGGCCGGAGAC- -......((((((.(((((((...(((.(((..(((((((...))))).)).((....))))).)))....))))))--).((....)).)))))).....- ( -31.80, z-score = -1.68, R) >droYak2.chrX 19821483 98 + 21770863 -CAAAGGCGGCCAAGUGGAAGAGCGUCGCAGAAGUGCCAUAAAAUGGCGACACCAAAUGGCUGCGAUUUAAUUUCCA--CACUCGAGGGCAGGCCGGAGAC- -......(((((..(((((((...(((((((..(((((((...))))).)).((....)))))))))....))))))--).((....))..))))).....- ( -36.60, z-score = -3.10, R) >droSec1.super_8 2830918 98 + 3762037 -CAAAGGCGGCCAAGUGGAAGAGCGUCACAGAAGUGCCAUAAAAUGGCGACACCAAAUGGCUGCGAUUUAAUUUCCA--CACUCGAGGGCUGGCCGGAGAC- -......((((((.(((((((...(((.(((..(((((((...))))).)).((....))))).)))....))))))--).((....)).)))))).....- ( -31.80, z-score = -1.68, R) >dp4.chrXL_group1e 9235640 85 + 12523060 CGAGAGGCAGCCAAGUGGAAAAGCGUCACAAAAGUGCCAUAAAAUGGCAACACCAAAUGGCUGCGAUUUAAUUUCCA--CUGUUGAG--------------- (((.(((((((((..(((....((.........))(((((...)))))....)))..)))))))((.......))..--)).)))..--------------- ( -25.40, z-score = -2.46, R) >droPer1.super_22 719031 85 - 1688296 CGAGAGGCAGCCAAGUGGAAAAGCGUCACAAAAGUGCCAUAAAAUGGCAACACCAAAUGGCUGCGAUUUAAUUUCCA--CUGUUGAG--------------- (((.(((((((((..(((....((.........))(((((...)))))....)))..)))))))((.......))..--)).)))..--------------- ( -25.40, z-score = -2.46, R) >droSim1.chrX 15816156 98 + 17042790 -CAAAGGCGGCCAAGUGGAAGAGCGUCACAGAAGUGCCAUAAAAUGGCGACACCAAAUGGCUGCGAUUUAAUUUCCA--CACUCGAGGGCUGGCCGAAGAC- -......((((((.(((((((...(((.(((..(((((((...))))).)).((....))))).)))....))))))--).((....)).)))))).....- ( -31.60, z-score = -1.93, R) >droAna3.scaffold_12903 697955 95 - 802071 -GGAGGGCAGCCAAGUGGAAAAGCGUCACAAAAGUGCCAUAAAAUGGCGACACCAAAUGGCUGCGAUUUAAUUUCCA------CACUAGGUUGCGAUGGGAA -.....(((((((..(((......(((((....))(((((...)))))))).)))..))))))).......((((((------(.(......).).)))))) ( -27.60, z-score = -1.18, R) >droWil1.scaffold_180702 1828804 101 - 4511350 -GGAGUUUAGCCAAGUGGAAAAGCAACGCAAAAGUGCCAUAAAAUGGCGACACCAAAUGGCUGCGAUUUAAUUUCCAAGUGCCCGCCAGCUGGUGACAACAA -.(((((((((((..(((....((...))....(((((((...))))).)).)))..)))))).))))).........((...((((....))))...)).. ( -26.30, z-score = -0.26, R) >droVir3.scaffold_12970 4914672 95 + 11907090 -CGACGGCAGCCAAGUGGAAGUGGAUUAGAAGAGUGCCAUAAAAUGGCGACACCAAAUGCUAGCGAUCUAA-UUCAA--CGGCAGGCGGUGCGCAGGAA--- -.....((((((..((.(...(((((((((...(((((((...))))).))......((....)).)))))-)))).--).)).)))..))).......--- ( -23.00, z-score = 0.54, R) >droGri2.scaffold_15081 3849178 93 - 4274704 -UGUGGGCUGCCAAGUGAAAGUGGCGAGCAAAAGUGCCGUAAAAUGUCGACGCCAAAUGCUUGCGAUUUA--UUUCA----CUGGCGAGUGUACGGGGAA-- -.(((.(((((((.((((((((.(((((((...(((.((........)).)))....))))))).))...--)))))----))))).))).)))......-- ( -34.10, z-score = -2.46, R) >consensus _CAAAGGCAGCCAAGUGGAAGAGCGUCACAAAAGUGCCAUAAAAUGGCGACACCAAAUGGCUGCGAUUUAAUUUCCA__CACUCGAGGGCUGGCCGGAGAC_ ......(((((((..(((.........((....))(((((...)))))....)))..)))))))...................................... (-18.64 = -18.69 + 0.05)
| Location | 20,594,058 – 20,594,156 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.96 |
| Shannon entropy | 0.52764 |
| G+C content | 0.51026 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -13.17 |
| Energy contribution | -12.91 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.863496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
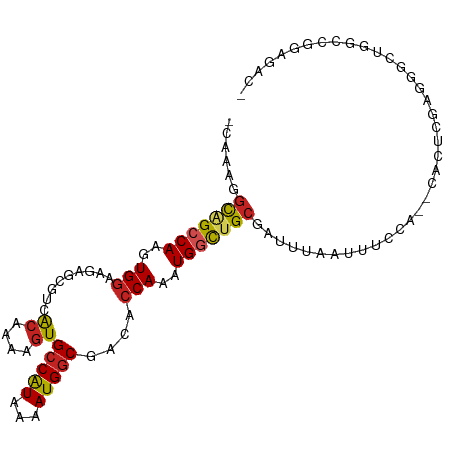
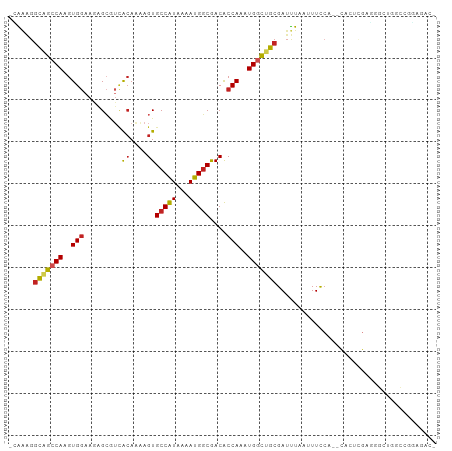
>dm3.chrX 20594058 98 - 22422827 -GUCUCCGGCCAGCCCUCGAGUG--UGGAAAUUAAAUCGCAGCCAUUUGGUGUCGCCAUUUUAUGGCACUUCUGUGACGCUCUUCCACUUGGCCGCCUUUG- -.....((((((((......))(--(((((......((((((((((.(((.....)))....))))).....))))).....)))))).))))))......- ( -29.80, z-score = -1.61, R) >droEre2.scaffold_4690 17189425 98 - 18748788 -GUCUCCGGCCAGCCCUCGAGUG--UGGAAAUUAAAUCGCAGCCAUUUGGUGUCGCCAUUUUAUGGCACUUCUGUGACGCUCUUCCACUUGGCCGCCUUUG- -.....((((((((......))(--(((((......((((((((((.(((.....)))....))))).....))))).....)))))).))))))......- ( -29.80, z-score = -1.61, R) >droYak2.chrX 19821483 98 - 21770863 -GUCUCCGGCCUGCCCUCGAGUG--UGGAAAUUAAAUCGCAGCCAUUUGGUGUCGCCAUUUUAUGGCACUUCUGCGACGCUCUUCCACUUGGCCGCCUUUG- -.....(((((.((......))(--(((((......((((((((((.(((.....)))....))))).....))))).....))))))..)))))......- ( -30.20, z-score = -1.75, R) >droSec1.super_8 2830918 98 - 3762037 -GUCUCCGGCCAGCCCUCGAGUG--UGGAAAUUAAAUCGCAGCCAUUUGGUGUCGCCAUUUUAUGGCACUUCUGUGACGCUCUUCCACUUGGCCGCCUUUG- -.....((((((((......))(--(((((......((((((((((.(((.....)))....))))).....))))).....)))))).))))))......- ( -29.80, z-score = -1.61, R) >dp4.chrXL_group1e 9235640 85 - 12523060 ---------------CUCAACAG--UGGAAAUUAAAUCGCAGCCAUUUGGUGUUGCCAUUUUAUGGCACUUUUGUGACGCUUUUCCACUUGGCUGCCUCUCG ---------------.....(((--((((((.....((((((((....)))..((((((...))))))....)))))....))))))).))........... ( -23.20, z-score = -1.89, R) >droPer1.super_22 719031 85 + 1688296 ---------------CUCAACAG--UGGAAAUUAAAUCGCAGCCAUUUGGUGUUGCCAUUUUAUGGCACUUUUGUGACGCUUUUCCACUUGGCUGCCUCUCG ---------------.....(((--((((((.....((((((((....)))..((((((...))))))....)))))....))))))).))........... ( -23.20, z-score = -1.89, R) >droSim1.chrX 15816156 98 - 17042790 -GUCUUCGGCCAGCCCUCGAGUG--UGGAAAUUAAAUCGCAGCCAUUUGGUGUCGCCAUUUUAUGGCACUUCUGUGACGCUCUUCCACUUGGCCGCCUUUG- -.....((((((((......))(--(((((......((((((((((.(((.....)))....))))).....))))).....)))))).))))))......- ( -29.60, z-score = -1.60, R) >droAna3.scaffold_12903 697955 95 + 802071 UUCCCAUCGCAACCUAGUG------UGGAAAUUAAAUCGCAGCCAUUUGGUGUCGCCAUUUUAUGGCACUUUUGUGACGCUUUUCCACUUGGCUGCCCUCC- ...(((.(((......)))------)))..........(((((((..((((((((((((...)))))((....)))))).....)))..))))))).....- ( -30.10, z-score = -3.36, R) >droWil1.scaffold_180702 1828804 101 + 4511350 UUGUUGUCACCAGCUGGCGGGCACUUGGAAAUUAAAUCGCAGCCAUUUGGUGUCGCCAUUUUAUGGCACUUUUGCGUUGCUUUUCCACUUGGCUAAACUCC- ..(((((((.....))))((.((..((((((...((.(((((((((.(((.....)))....))))).....))))))...))))))..)).)).)))...- ( -26.50, z-score = -0.18, R) >droVir3.scaffold_12970 4914672 95 - 11907090 ---UUCCUGCGCACCGCCUGCCG--UUGAA-UUAGAUCGCUAGCAUUUGGUGUCGCCAUUUUAUGGCACUCUUCUAAUCCACUUCCACUUGGCUGCCGUCG- ---.....((((((((..(((..--.(((.-.....)))...)))..))))).)))......((((((..(...................)..))))))..- ( -20.61, z-score = -0.16, R) >droGri2.scaffold_15081 3849178 93 + 4274704 --UUCCCCGUACACUCGCCAG----UGAAA--UAAAUCGCAAGCAUUUGGCGUCGACAUUUUACGGCACUUUUGCUCGCCACUUUCACUUGGCAGCCCACA- --..............(((((----(((((--......((.((((...((.((((........)))).))..)))).))...)))))).))))........- ( -25.00, z-score = -2.39, R) >consensus _GUCUCCGGCCAGCCCUCGAGUG__UGGAAAUUAAAUCGCAGCCAUUUGGUGUCGCCAUUUUAUGGCACUUUUGUGACGCUCUUCCACUUGGCCGCCUUUG_ .........................(((((......((((((......(((((((........))))))).)))))).....)))))............... (-13.17 = -12.91 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:32 2011