| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,593,416 – 20,593,516 |
| Length | 100 |
| Max. P | 0.845988 |

| Location | 20,593,416 – 20,593,516 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 71.97 |
| Shannon entropy | 0.56573 |
| G+C content | 0.40660 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -11.71 |
| Energy contribution | -12.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
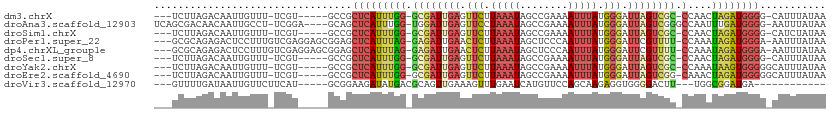
>dm3.chrX 20593416 100 + 22422827 ---UCUUAGACAAUUGUUU-UCGU-----GCCGCUCAUUUGG-GCGAUUGAGUUCUUAAAUAGCCGAAAAUUUAUGGGAUUAGUCGC-CCAACUAGAUGGGG-CAUUUAUAA ---....((((....))))-..((-----(((.(((..((((-((((((((.(((.(((((........))))).))).))))))))-))))...)).).))-)))...... ( -30.00, z-score = -2.45, R) >droAna3.scaffold_12903 697471 105 - 802071 UCAGCGACAACAAUUGCCU-UCGGA----GCAGCUCAUUUGG-UGGAUUGAGUUCCUAAAUAGCCGAAAAUUUAUGGGAUUAGUCGGGCCAAUUUGAUGGGG-AAUUUAUAA .............(((((.-...).----))))(((((((((-(.((((((.(((((((((........))))).)))))))))).).)))....)))))).-......... ( -22.70, z-score = 0.70, R) >droSim1.chrX 15815528 100 + 17042790 ---UCUUAGACAAUUGUUU-UCGU-----GCCGCUCAUUUGG-GCGAUUGAGUUCUUAAAUAGCCGAAAAUUUAUGGGAUUAGUCGC-CCAACUAGAUGGGG-CAUCUAUAA ---....((((....))))-..((-----(((.(((..((((-((((((((.(((.(((((........))))).))).))))))))-))))...)).).))-)))...... ( -30.00, z-score = -2.28, R) >droPer1.super_22 718206 106 - 1688296 ---GCGCAGAGACUCCUUUGUCGAGGAGCGGAGCUCAUUUAG-GAGAUUGAACUCUUAAAUAGCUCCCAAUUUAUGGGAUUCGUUUU-CCAAAUAGAUGGGA-AAUUUAUAA ---.(.((..(.((((((....)))))))((((((.((((((-(((......))))))))))))))).......)).)....(((((-(((......)))))-)))...... ( -33.50, z-score = -2.67, R) >dp4.chrXL_group1e 9234878 106 + 12523060 ---GCGCAGAGACUCCUUUGUCGAGGAGCGGAGCUCAUUUAG-GAGAUUGAACUCUUAAAUAGCUCCCAAUUUAUGGGAUUCGUUUU-CCAAAUAGAUGGGA-AAUUUAUAA ---.(.((..(.((((((....)))))))((((((.((((((-(((......))))))))))))))).......)).)....(((((-(((......)))))-)))...... ( -33.50, z-score = -2.67, R) >droSec1.super_8 2830290 100 + 3762037 ---UCUUAGACAAUUGUUU-UCGU-----GCCGCUCAUUUGG-GCGAUUGAGUUCUUAAAUAGCCGAAAAUUUAUGGGAUUAGUCGC-CCAACUAGAUGGGG-CAUUUAUAA ---....((((....))))-..((-----(((.(((..((((-((((((((.(((.(((((........))))).))).))))))))-))))...)).).))-)))...... ( -30.00, z-score = -2.45, R) >droYak2.chrX 19820832 101 + 21770863 ---UCUUAGACAAUUGUUU-UCGU-----GCCGCUCAUUUGG-GCGAUUGAGUUCUUAAAUAGCCGAAAAUUUAUGGGAUUAGUCGC-CCAAAUAAGUGGGGGCAUUUAUAA ---....((((....))))-..((-----(((.(((((((((-((((((((.(((.(((((........))))).))).))))))))-))))....))))))))))...... ( -32.60, z-score = -3.11, R) >droEre2.scaffold_4690 17188835 101 + 18748788 ---UCUUAGACAAUUGUUU-UCGU-----GCCGCUCAUUUGG-GCGAUUGAGUUCUUAAAUAGCCGAAAAUUUAUGGGAUUAGUCGG-CAAACUAGAUGGGGGCAUUUAUAA ---....((((....))))-..((-----(((.(((((((((-..((......)).......(((((.((((.....))))..))))-)...))))))))))))))...... ( -26.90, z-score = -1.64, R) >droVir3.scaffold_12970 4913379 89 + 11907090 ---GUUUUGAUAAUUGUUCUUCAU-----GCGGAAGAUAUGACGCAGUUGAAAGUUUGAAUCAUGUUCCAGCAAGAGGUGGGGACUU---UGGCGGAUGA------------ ---.......(((((((...((((-----((....).))))).)))))))...(((((......(((((.((.....)).)))))..---...)))))..------------ ( -19.20, z-score = 0.30, R) >consensus ___UCUUAGACAAUUGUUU_UCGU_____GCCGCUCAUUUGG_GCGAUUGAGUUCUUAAAUAGCCGAAAAUUUAUGGGAUUAGUCGC_CCAACUAGAUGGGG_CAUUUAUAA .................................((((((((..((((((((.(((.(((((........))))).))).))))))))......))))))))........... (-11.71 = -12.14 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:30 2011