| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,554,543 – 20,554,609 |
| Length | 66 |
| Max. P | 0.925198 |

| Location | 20,554,543 – 20,554,609 |
|---|---|
| Length | 66 |
| Sequences | 9 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 72.05 |
| Shannon entropy | 0.57909 |
| G+C content | 0.42537 |
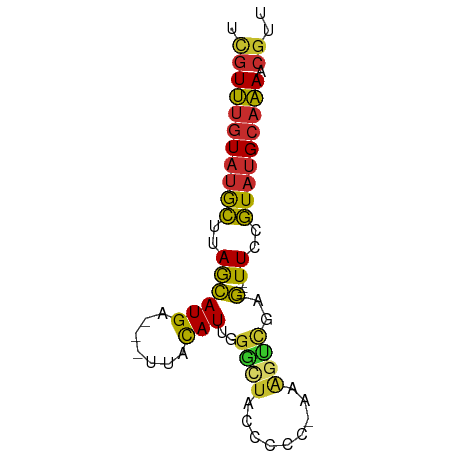
| Mean single sequence MFE | -18.08 |
| Consensus MFE | -7.02 |
| Energy contribution | -6.91 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.925198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
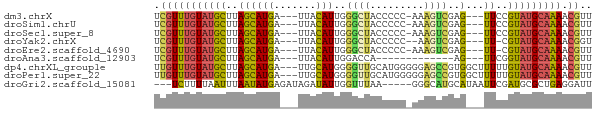
>dm3.chrX 20554543 66 - 22422827 UCGUUUGUAUGCUUAGCAUGA---UUACAUUGGGCUACCCCC-AAAGUCGAG---UUCCGUAUGCAAAACGUU .(((((((((((..(((.(((---((...(((((.....)))-))))))).)---))..)))))).))))).. ( -20.00, z-score = -3.18, R) >droSim1.chrU 14684363 66 - 15797150 UCGUUUGUAUGCUUAGCAUGA---UUACAUUGGGCUACCCCC-AAAGUCGAG---UUCCGUAUGCAAAACGUU .(((((((((((..(((.(((---((...(((((.....)))-))))))).)---))..)))))).))))).. ( -20.00, z-score = -3.18, R) >droSec1.super_8 2792213 66 - 3762037 UCGUUUGUAUGCUUAGCAUGA---UUACAUUGGGCUACCCCC-AAAGUCGAG---UUCCGUAUGCAAAACGUU .(((((((((((..(((.(((---((...(((((.....)))-))))))).)---))..)))))).))))).. ( -20.00, z-score = -3.18, R) >droYak2.chrX 19781560 64 - 21770863 UCGUUUGUAUGCUUAGCAUGA---UUACAUUGGGCUACCCCC--AAGUCGAG---UU-CGUAUGCAAAACGGU ((((((((((((..(((.(((---(....(((((.....)))--)))))).)---))-.)))))).)))))). ( -18.70, z-score = -2.08, R) >droEre2.scaffold_4690 17150009 65 - 18748788 UCGUUUGUAUGCUUAGCAUGA---UUACAUUGGGCUACCCCC-AAAGUCGAG---UU-CGUAUGCAAAACGUU .(((((((((((..(((.(((---((...(((((.....)))-))))))).)---))-.)))))).))))).. ( -18.80, z-score = -2.73, R) >droAna3.scaffold_12903 641506 54 + 802071 UCGUUUGUAUGCUUAGCAUGA---UUACAUUGGACCA-------------AG---UUCGGUAUGCAAAACGUU .((((((((((((.(((.((.---((......)).))-------------.)---)).))))))).))))).. ( -13.40, z-score = -1.63, R) >dp4.chrXL_group1e 9190396 70 - 12523060 UUGUUUGUAUGCUUAGCAUGA---UUGCAUGGGGUUGCAUGGGGGAGCCGUGGCUUUUUGUAUGCAAAACGUU .(((...(((((...))))).---..)))..(..((((((((((((((....))))))).)))))))..)... ( -20.90, z-score = -1.31, R) >droPer1.super_22 672495 70 + 1688296 UUGUUUGUAUGCUUAGCAUGA---UUGCAUGGGGUUGCAUGGGGGAGCCGUGGCUUUUUGUAUGCAAAACGUU .(((...(((((...))))).---..)))..(..((((((((((((((....))))))).)))))))..)... ( -20.90, z-score = -1.31, R) >droGri2.scaffold_15081 3802156 65 + 4274704 ---UCUUUUAAUUUAAUAUGAGAUAGAUAUUGGUUUAA-----GGGCAUGCAUAAUUCGAUGCGCUGAGGAUU ---.(((((((.(((((((.......))))))).))))-----)))(((((((......))))).))...... ( -10.00, z-score = -0.05, R) >consensus UCGUUUGUAUGCUUAGCAUGA___UUACAUUGGGCUACCCCC_AAAGUCGAG___UUCCGUAUGCAAAACGUU .(((((((((((.....(((.......)))..((((.........))))..........))))))))).)).. ( -7.02 = -6.91 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:27 2011