| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,183,465 – 12,183,560 |
| Length | 95 |
| Max. P | 0.944633 |

| Location | 12,183,465 – 12,183,560 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.63 |
| Shannon entropy | 0.16328 |
| G+C content | 0.40598 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.49 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
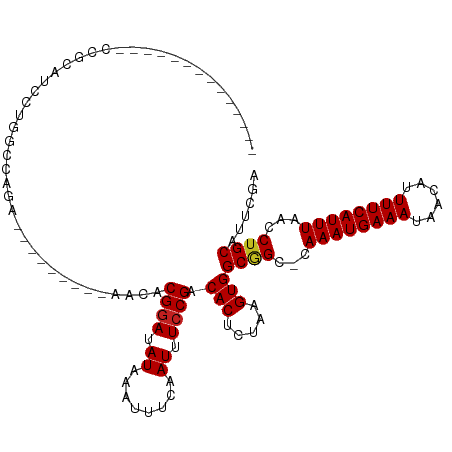
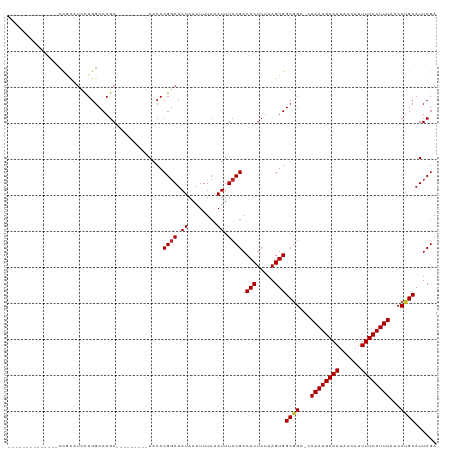
>dm3.chr2L 12183465 95 + 23011544 --------------CCGCAUCCUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGC-CAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------..(((....((((...---------....((((.((........)).)))).(((.....)))..)))-)((((((((......))))))))....)))...... ( -24.00, z-score = -3.16, R) >droSim1.chr2L 11984250 95 + 22036055 --------------CCGCAUCCUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGC-CAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------..(((....((((...---------....((((.((........)).)))).(((.....)))..)))-)((((((((......))))))))....)))...... ( -24.00, z-score = -3.16, R) >droSec1.super_16 378960 95 + 1878335 --------------CCGCAUCCUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGC-CAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------..(((....((((...---------....((((.((........)).)))).(((.....)))..)))-)((((((((......))))))))....)))...... ( -24.00, z-score = -3.16, R) >droYak2.chr2L 8613636 95 + 22324452 --------------CCGCAUCCUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGC-CAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------..(((....((((...---------....((((.((........)).)))).(((.....)))..)))-)((((((((......))))))))....)))...... ( -24.00, z-score = -3.16, R) >droEre2.scaffold_4929 13412183 95 - 26641161 --------------CCGCAUCCUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGC-CAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------..(((....((((...---------....((((.((........)).)))).(((.....)))..)))-)((((((((......))))))))....)))...... ( -24.00, z-score = -3.16, R) >droAna3.scaffold_12916 2981315 95 - 16180835 --------------CCGCAUCCUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGC-CAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------..(((....((((...---------....((((.((........)).)))).(((.....)))..)))-)((((((((......))))))))....)))...... ( -24.00, z-score = -3.16, R) >dp4.chr4_group3 5711827 95 - 11692001 --------------CCACAUCCUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGC-CAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------.........((((...---------....((((.((........)).)))).(((.....)))..)))-)((((((((......))))))))............. ( -20.90, z-score = -2.45, R) >droPer1.super_1 2807810 95 - 10282868 --------------CCACAUCCUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGC-CAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------.........((((...---------....((((.((........)).)))).(((.....)))..)))-)((((((((......))))))))............. ( -20.90, z-score = -2.45, R) >droWil1.scaffold_180708 2699806 118 - 12563649 CCACAUACACAGAUAUCCUUUCUGGCCAGAGAAGACAGAAACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGC-GGGCAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA ............((((((((((((...........))))))...)))))).............((((((.....)))((-(((.((((((((......))))))))..)))))..))). ( -28.00, z-score = -3.38, R) >droVir3.scaffold_12963 17244665 96 + 20206255 --------------CUGCUGCCUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGGGCAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------.(((.((((.(((...---------....((((.((........)).))))..((.....))))).))))((((((((......)))))))).....)))..... ( -26.00, z-score = -2.82, R) >droGri2.scaffold_15252 14673967 96 + 17193109 ---------------CCUCUUCUGGCCAGA--------AAACACGCAUAUAAAUUUCAAUUUCCGACACUCGAAGUGGCAGGGCAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA ---------------....((((....)))--------)..............................(((((...(((((..((((((((......))))))))..))))).))))) ( -23.60, z-score = -3.12, R) >droMoj3.scaffold_6500 22285935 96 + 32352404 --------------CUGCUGCUUGGCCAGA---------AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGGGCAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA --------------..(((....)))..((---------(...((((.((........)).)))).(((.....)))(((((..((((((((......))))))))..))))).))).. ( -25.30, z-score = -2.78, R) >consensus ______________CCGCAUCCUGGCCAGA_________AACACGGAUAUAAAUUUCAAUUUCCGACACUCUAAGUGGCGGC_CAAAUGAAAUAACAUUUUCAUUUAACCUGCAUUCGA ...........................................((((.((........)).)))).(((.....)))((((...((((((((......))))))))...))))...... (-16.39 = -16.49 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:31 2011