
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,531,052 – 20,531,144 |
| Length | 92 |
| Max. P | 0.545606 |

| Location | 20,531,052 – 20,531,144 |
|---|---|
| Length | 92 |
| Sequences | 14 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 72.71 |
| Shannon entropy | 0.62294 |
| G+C content | 0.50972 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -11.72 |
| Energy contribution | -12.51 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20531052 92 + 22422827 UUUCGGCC---GAAUGUGCGUCGAAUAAUCGACUUGUUUGUGAUCUUUUACCUGCCAAAACUGGUGCCGCUGCUGCGGCCC-AAACUUUUCACGGA ......((---(..((.(((((((....)))))......((((....))))..))))....(((.(((((....)))))))-).........))). ( -27.30, z-score = -1.84, R) >droWil1.scaffold_180702 1761967 92 - 4511350 UUUCGUCC---CAACAUGCGACGAAUUAUUGACUUUUUUGUGAUAUUCUAUUUGCCCAAAUUGGUGCCGCUGCUGCGCUCG-AAACUCUUCACCGA .((((((.---(.....).))))))..............((((..(((.....((((.....)).))(((....)))...)-)).....))))... ( -15.70, z-score = -0.23, R) >droSim1.chrX 15754950 92 + 17042790 UUUCGGCC---GAAUGUGCGUCGAGUAAUCGACUUGUUUGUGAUCUUUUACCUGCCCAAACUGGUGCCGCUGCUGCGGCCC-AAACUUUUCACGGA ......((---(..((.(((((((....)))))......((((....))))..)).))...(((.(((((....)))))))-).........))). ( -29.60, z-score = -2.13, R) >droSec1.super_8 2773809 92 + 3762037 UUUCGGCC---GAAUGUGCGUCGAGUAAUCGACUUGUUUCUGAUCUUUUACCUGCCAAAACUGGUGCCGCUGCUGCGGCCC-AAACUUUUCACGGA ......((---(..((.(((((((....))))).......(((....)))...))))....(((.(((((....)))))))-).........))). ( -26.70, z-score = -1.44, R) >droYak2.chrX 19763116 92 + 21770863 UUUCGGCC---GAAUGUGCGUCGAGUAAUCGACUUGUUUGUGAUCUUUUACCUGCCAAAACUGGUGCCGCUGCUGCGGCCA-AAACUUUUCACGGA ......((---........(((((....)))))......((((..........((((....))))(((((....)))))..-.......)))))). ( -26.90, z-score = -1.22, R) >droEre2.scaffold_4690 17131446 92 + 18748788 UUUCGGCC---GAAUGUGCGUCGAGUAAUCGACUUGUUUGUGAUCUUUUACCUGCCAAAACUGGUGCCGCUGCUGCGGCCC-AAACUUUUCACGGA ......((---(..((.(((((((....)))))......((((....))))..))))....(((.(((((....)))))))-).........))). ( -29.60, z-score = -2.12, R) >droAna3.scaffold_12903 620701 92 - 802071 UUCAAGCC---GAAUGUUUGGCGGAUAAUCGACUUGUUUGUGAUCUUCUACCUGCCAAAAUUUGUGCCGCUAUUGCGGCCG-AAACUUUUUACGGA ......((---(....(((((((((..(((.((......)))))..)))....)))))).((((.(((((....)))))))-))........))). ( -27.50, z-score = -2.61, R) >droVir3.scaffold_12970 4833731 92 + 11907090 UUUAGGCC---GAAUGUGCGCCGUAUAAUAGACUUCUUUGUGAUAUUCUUUCUGCCCAAGCUGGUGCCGCUGUUGCGGUCC-AAGCUGUUUACGGA ....((((---(....)).)))((((.(((((....))))).))))....((((....((((((.(((((....)))))))-.)))).....)))) ( -23.10, z-score = 0.19, R) >droMoj3.scaffold_6473 4306635 93 + 16943266 UUCAAGCC---GAAUGUGCGUCGAAUAAUAGACUUCUUUGUGAUAUUCUUUCUGCCGAAGCUGGUGCCGCUGAUGCGGUUCGAAGCUGUUCACGGA ........---(((((((((..(((........)))..))).))))))......(((.((((...(((((....)))))....)))).....))). ( -23.10, z-score = 0.46, R) >droGri2.scaffold_15081 3784854 92 - 4274704 UUUCGACC---GAAUGUGCGUCGCAUGGUUGACUUCUUUGUGAUAUUCUUUCUGCCCAAGCUGGCGCCGCUGUUACGGUGC-AAGCUGUUCACAGA ..((((((---..(((((...)))))))))))....(((((((...............((((.((((((......))))))-.))))..))))))) ( -28.73, z-score = -1.15, R) >dp4.chrXL_group1e 9173916 92 + 12523060 UUCCGGCC---GAAUGUGCGGCGGAUAAUUGACUUUUUCGUGAUAUUUUACCUGCCCAAGCUGGUGCCGCUGUUGCGGCCC-CGUCUGUUCACGGA .(((((((---(......))))(((((...(((......((((....))))...........((.(((((....))))).)-))))))))).)))) ( -31.20, z-score = -1.27, R) >droPer1.super_22 654914 92 - 1688296 UUCCGGCC---GAAUGUGCGGCGGAUAAUUGACUUUUUCGUGAUAUUUUACCUGCCCAAGCUGGUGCCGCUGUUGCGGCCC-CGUCUGUUCACGGA .(((((((---(......))))(((((...(((......((((....))))...........((.(((((....))))).)-))))))))).)))) ( -31.20, z-score = -1.27, R) >anoGam1.chr3L 28506383 89 - 41284009 -UUCGACUACGGACCGGGGCGCGGCCUGACGAUGGCAUCGU--UCUUCUUUCUGCCCGAGCUGGUGCCUUAUUUGCGGCUG-AAGCUUUUCCC--- -.........(((..(((((.(((((.(.(((.((((((((--((............)))).))))))....)))))))))-..)))))))).--- ( -26.80, z-score = 0.60, R) >apiMel3.Group14 4628865 76 + 8318479 ------------UCCCAGCUGUCGACGUUCAAGCAGAAUUUGAUGAUAUUCGUGCACAGGAACUCGCCCCGUCU--GGCCA--GAUUGUUCA---- ------------..((((.((.(((.((((..(((((((........)))).)))....)))))))...)).))--))...--.........---- ( -15.90, z-score = 0.63, R) >consensus UUUCGGCC___GAAUGUGCGUCGAAUAAUCGACUUGUUUGUGAUCUUUUACCUGCCCAAACUGGUGCCGCUGCUGCGGCCC_AAACUUUUCACGGA ....................................(((((((..........(((......)))(((((....)))))..........))))))) (-11.72 = -12.51 + 0.79)

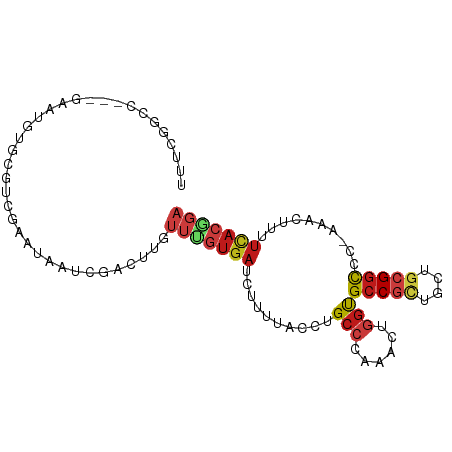

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:23 2011