| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,524,919 – 20,524,992 |
| Length | 73 |
| Max. P | 0.823072 |

| Location | 20,524,919 – 20,524,992 |
|---|---|
| Length | 73 |
| Sequences | 12 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 84.01 |
| Shannon entropy | 0.32380 |
| G+C content | 0.34003 |
| Mean single sequence MFE | -8.47 |
| Consensus MFE | -8.06 |
| Energy contribution | -7.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823072 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
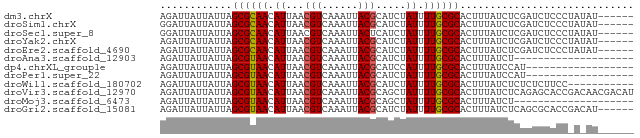
>dm3.chrX 20524919 73 + 22422827 AGAUUAUUAUUAGCGCAACAUUAACGUCAAAUUACGCAUCUAUUUUGCGCACUUUAUCUCGAUCUCCCUAUAU------ ............((((((.((...(((......))).....)).)))))).......................------ ( -9.40, z-score = -1.30, R) >droSim1.chrX 15748603 73 + 17042790 GGAUUAUUAUUAGCGCAACAUUAACGUCAAAUUACGCAUCUAUUUUGCGCACUUUAUCUCGAUCUCCCUAUAU------ ............((((((.((...(((......))).....)).)))))).......................------ ( -9.40, z-score = -1.06, R) >droSec1.super_8 2767612 73 + 3762037 GGAUUAUUAUUAGCGCAACAUUAACGUCAAAUUACUCAUCUAUUUUGCGCACUUUAUCUCGAUCUCCCUAUAU------ ............((((((.((....................)).)))))).......................------ ( -7.05, z-score = -0.72, R) >droYak2.chrX 19755280 73 + 21770863 AGAUUAUUAUUAGCGCAACAUUAACGUCAAAUUACGCAUCUAUUUUGCGCACUUUAUCUCGAUCUCCCUAUAU------ ............((((((.((...(((......))).....)).)))))).......................------ ( -9.40, z-score = -1.30, R) >droEre2.scaffold_4690 17125335 73 + 18748788 AGAUUAUUAUUAGCGCAACAUUAACGUCAAAUUACGCAUCUAUUUUGCGCACUUUAUCUCGAUCUCCCUAUAU------ ............((((((.((...(((......))).....)).)))))).......................------ ( -9.40, z-score = -1.30, R) >droAna3.scaffold_12903 613888 59 - 802071 AGAUUAUUAUUAGCGUAACAUUAACGUCAAAUUACGCAUCUAUUUUGCGCACUUUAUCU-------------------- ............((((((.((...(((......))).....)).)))))).........-------------------- ( -7.40, z-score = -0.93, R) >dp4.chrXL_group1e 9168084 61 + 12523060 AGAUUAUUAUUAGCGUAACAUUAACGUCAAAUUACGCAUCCAUUUUGCGCACUUUAUCCAU------------------ ............((((((......(((......)))........))))))...........------------------ ( -6.74, z-score = -1.04, R) >droPer1.super_22 649080 61 - 1688296 AGAUUAUUAUUAGCGUAACAUUAACGUCAAAUUACGCAUCUAUUUUGCGCACUUUAUCCAU------------------ ............((((((.((...(((......))).....)).))))))...........------------------ ( -7.40, z-score = -1.08, R) >droWil1.scaffold_180702 1752208 68 - 4511350 AGAUUAUUAUUAGCGUAACAUUAACGUCAAAUUACGCAUCUAUUUUGCGCACUUUAUCUCUCUCUUCC----------- ............((((((.((...(((......))).....)).))))))..................----------- ( -7.40, z-score = -1.29, R) >droVir3.scaffold_12970 4827584 79 + 11907090 AGAUUAUUAUUAGCGUAACAUUAACGUCAAAUUACGCAGCUAUUUUGCGCACUUUAUCUCAGAGCACCGACAACGACAU ............((((((.............)))))).((......))((.((.......)).)).............. ( -8.62, z-score = 0.61, R) >droMoj3.scaffold_6473 4289320 59 + 16943266 AGAUUAUUAUUAGCGUAACAUUAACGUCAAAUUACGCAGCUAUUUUGCGCACUUUAUCU-------------------- ((((........((((((.............)))))).((......)).......))))-------------------- ( -7.62, z-score = -0.28, R) >droGri2.scaffold_15081 3779588 73 - 4274704 AGAUUAUUAUUAGCGUAACAUUAACGUCAAAUUACGCAUCUAUUUUGCGCACUUUAUCUCAGCGCACCGACAU------ ............((((((.............))))))........(((((...........))))).......------ ( -11.82, z-score = -1.28, R) >consensus AGAUUAUUAUUAGCGUAACAUUAACGUCAAAUUACGCAUCUAUUUUGCGCACUUUAUCUCGAUCUCCC_A_AU______ ............((((((.((...(((......))).....)).))))))............................. ( -8.06 = -7.90 + -0.16)

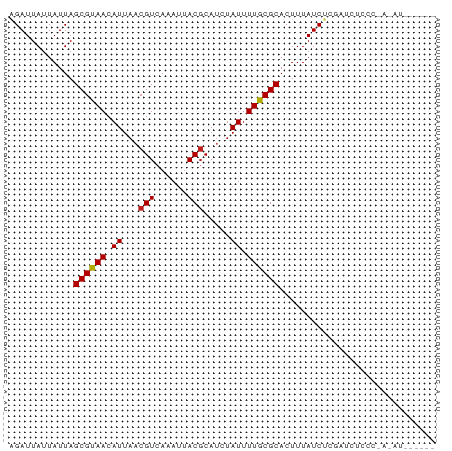
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:22 2011