
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,492,109 – 20,492,201 |
| Length | 92 |
| Max. P | 0.761973 |

| Location | 20,492,109 – 20,492,201 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 88.12 |
| Shannon entropy | 0.22966 |
| G+C content | 0.42121 |
| Mean single sequence MFE | -13.46 |
| Consensus MFE | -11.26 |
| Energy contribution | -11.26 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20492109 92 + 22422827 GUUUAUUUCAUUUUAUUAUUUUUUCUCUUUUGCGGUCAACGAGUUGGUUGACCACUUUCAAAUACCGCCCACGCCCGU-CCCUCAACGCACCA------- ..............................((((((((((......))))))).............((....))....-........)))...------- ( -14.20, z-score = -1.52, R) >droSim1.chrX 15713204 93 + 17042790 GUUUAUUUCAUUUUAUUAUUUUUUCUCUUUUGCGGUCAACGAGUUGGUUGACCACUUUCAAAUACCUCCCACGCCCCUUCCCUCAACGCACCG------- .................................(((((((......)))))))........................................------- ( -11.30, z-score = -1.31, R) >droSec1.super_8 2735145 93 + 3762037 GUUUAUUUCAUUUUAUUAUUUUUUCUCUUUUGCGGUCAACGAGUUGGUUGACCACUUUCAAAUACCGCCCACGCCCCUUCCCUCAACGCACCG------- ..............................((((((((((......))))))).............((....)).............)))...------- ( -14.20, z-score = -2.06, R) >droYak2.chrX 19722016 92 + 21770863 GUUUAUUUCAUUUUAUUAUUUUUUCUCUUUUGCGGUCAACGAGUUGGUUGACCACUUUCAAAUACCGCCCACGCCCCU-CCCUCAACGCACCG------- ..............................((((((((((......))))))).............((....))....-........)))...------- ( -14.20, z-score = -2.08, R) >droEre2.scaffold_4690 17093394 92 + 18748788 GUUUAUUUCAUUUUAUUAUUUUUUCUCUUUUGCGGUCAACGAGUUGGUUGACCACUUUCAAAUACCGCCCACGCCCGC-CCCUCAACGCACCA------- ..............................((((((((((......))))))).............((....))....-........)))...------- ( -14.20, z-score = -1.47, R) >droAna3.scaffold_12903 574039 100 - 802071 GUUUAUUUCAUUUUAUUAUUUUUUCUCUUUUGUGGUCAACGAGUUGGUUGACCACUUUCAAAUACAGCACACCAAUGGCCCGCCCUCCCUCUAUUUGCUU ...............................(((((((((......)))))))))..........((((.......((........)).......)))). ( -16.94, z-score = -1.65, R) >dp4.chrXL_group1e 9129599 92 + 12523060 GUUUAUUUCAUUUUAUUAUUUUUUCUCUUUUGCGGUCAACGAGUUGGUUGACCACUUUCAAAUACAGCCCACCGACCU-UUCUCCACACCCAA------- .................................(((((((......))))))).........................-..............------- ( -11.30, z-score = -1.06, R) >droPer1.super_22 611182 92 - 1688296 GUUUAUUUCAUUUUAUUAUUUUUUCUCUUUUGCGGUCAACGAGUUGGUUGACCACUUUCAAAUACAGCCCACCGACCU-UUCUCCACACCCAU------- .................................(((((((......))))))).........................-..............------- ( -11.30, z-score = -1.17, R) >consensus GUUUAUUUCAUUUUAUUAUUUUUUCUCUUUUGCGGUCAACGAGUUGGUUGACCACUUUCAAAUACCGCCCACGCCCCU_CCCUCAACGCACCA_______ .................................(((((((......)))))))............................................... (-11.26 = -11.26 + 0.00)

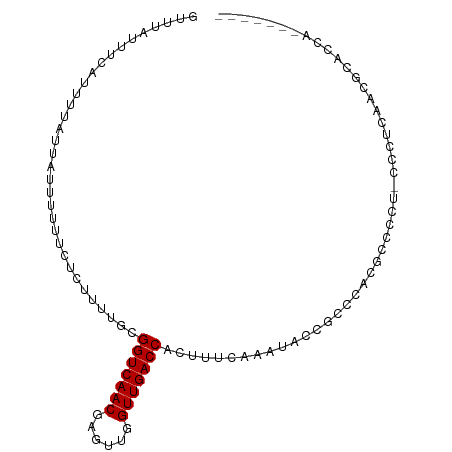
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:18 2011