
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,488,446 – 20,488,538 |
| Length | 92 |
| Max. P | 0.508634 |

| Location | 20,488,446 – 20,488,538 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Shannon entropy | 0.37452 |
| G+C content | 0.38146 |
| Mean single sequence MFE | -16.14 |
| Consensus MFE | -9.28 |
| Energy contribution | -8.92 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508634 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
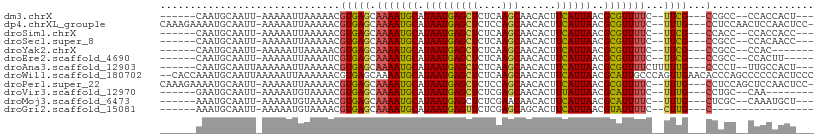
>dm3.chrX 20488446 92 - 22422827 ------CAAUGCAAUU-AAAAAUUAAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCAAGCAACACUUCAUUAACGCGUUUUC--UUCG---CCGCC--CCACCACU--- ------..........-.............(((((.(((((((.(((((((((....)))......))))))..))))))).--))))---)....--........--- ( -15.20, z-score = -1.55, R) >dp4.chrXL_group1e 9124859 102 - 12523060 CAAAGAAAAUGCAAUU-AAAAAUUAAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCCAGCAACACUUCAUUAACGCGUUUUC--UUUG---CCUCCAACUCCAACUCC- ((((((((((((....-.................((.....)).(((((((((....)))......))))))..))))))))--))))---.................- ( -20.10, z-score = -3.55, R) >droSim1.chrX 15708675 92 - 17042790 ------CAAUGCAAUU-AAAAAUUAAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCAAGCAACACUUCAUUAACGCGUUUUC--UUCG---CCACC--CCACCACC--- ------..........-.............(((((.(((((((.(((((((((....)))......))))))..))))))).--))))---)....--........--- ( -15.20, z-score = -2.34, R) >droSec1.super_8 2729979 92 - 3762037 ------CAAUGCAAUU-AAAAAUUAAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCAAGCAACACUUCAUUAACGCGUUUUC--UUCG---CCGCC--CCACAACC--- ------..........-.............(((((.(((((((.(((((((((....)))......))))))..))))))).--))))---)....--........--- ( -15.20, z-score = -1.57, R) >droYak2.chrX 19718331 88 - 21770863 ------CAAUGCAAUU-AAAAAUUAAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCAAGCAACACUUCAUUAACGCGUUUUC--UUCG---CCGCC--CCAC------- ------..........-.............(((((.(((((((.(((((((((....)))......))))))..))))))).--))))---)....--....------- ( -15.20, z-score = -1.46, R) >droEre2.scaffold_4690 17089614 90 - 18748788 ------CAAUGCAAUU-AAAAAUUAAAAUCGUGAGCAAAAUGCAUAAUGAGCUCUCAAGCAACACUUCAUUAACGCGUUUUC--UUCG---CCGCC--CCACUU----- ------..........-.............(((((.(((((((.(((((((((....)))......))))))..))))))).--))))---)....--......----- ( -15.20, z-score = -1.43, R) >droAna3.scaffold_12903 569940 95 + 802071 ------CAAUGCAAUUAAAAAAUUAAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCAAGCAACACUUCAUUAACGCGUUUUCUUUUUG---CCCCU--UUGCCACU--- ------....((((....((((...((((((((.((.....)).(((((((((....)))......)))))).))))))))..)))).---.....--))))....--- ( -17.10, z-score = -1.50, R) >droWil1.scaffold_180702 1711196 107 + 4511350 --CACCAAAUGCAAUUAAAAAUUAAAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCAAGCAACACUUCAUUAACGCAUUGCCCAGUUGAACACCCAGCCCCCCACUCCC --............................(((.(((..((((.(((((((((....)))......))))))..)))))))...((((......))))....))).... ( -14.20, z-score = -1.30, R) >droPer1.super_22 606637 102 + 1688296 CAAAGAAAAUGCAAUU-AAAAAUUAAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCCAGCAACACUUCAUUAACGCGUUUUC--UUUG---CCUCCAGCUCCAACUCC- ((((((((((((....-.................((.....)).(((((((((....)))......))))))..))))))))--))))---.................- ( -20.10, z-score = -2.82, R) >droVir3.scaffold_12970 4784036 87 - 11907090 ------GAAUGCAAUU-AAAAAUGUAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCGAGCAACACUUUAUUAACGCAUUUUC--UUUG---CCUGC--CAA-------- ------....((((..-.(((((((....(((..((.....))...))).(((....)))..............))))))).--.)))---)....--...-------- ( -13.60, z-score = 0.30, R) >droMoj3.scaffold_6473 4234083 92 - 16943266 ------AAAUGCAAUU-AAAAAUGUAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCGAACAACACUUCAUUAACGCAUUUUC--UUUG---CUCGC--CAAAUGCU--- ------...((((...-.....))))....(((((((((((((.(((((((..............)))))))..))))....--))))---)))))--........--- ( -18.44, z-score = -1.63, R) >droGri2.scaffold_15081 3736267 80 + 4274704 ------AAAUGCAAUU-AAAAAUGUAAAACGUGAGCAAAAUGCAUAAUGAGUUCUCGAGCAGCACUUCAUUAACGUAUUUUC--CUUG---C----------------- ------...((((...-.....))))....(..((.(((((((.((((((((.((.....)).)).))))))..))))))).--))..---)----------------- ( -14.20, z-score = -0.68, R) >consensus ______CAAUGCAAUU_AAAAAUUAAAAACGUGAGCAAAAUGCAUAAUGAGCUCUCAAGCAACACUUCAUUAACGCGUUUUC__UUUG___CCGCC__CCACCACU___ .......(((((......................((.....)).(((((((((....)))......))))))..))))).............................. ( -9.28 = -8.92 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:16 2011