| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,166,970 – 12,167,078 |
| Length | 108 |
| Max. P | 0.865107 |

| Location | 12,166,970 – 12,167,078 |
|---|---|
| Length | 108 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 64.86 |
| Shannon entropy | 0.81544 |
| G+C content | 0.46539 |
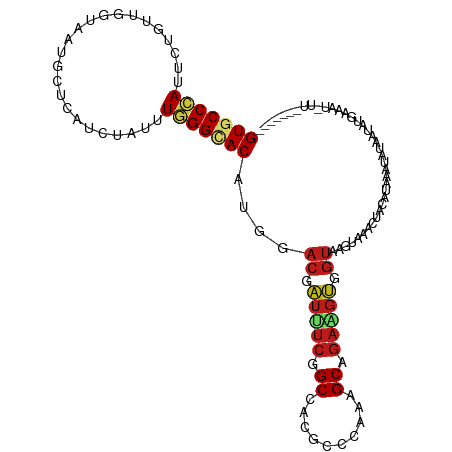
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -13.23 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.65 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.865107 |
| Prediction | RNA |
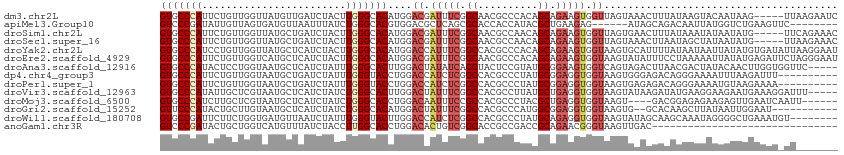
Download alignment: ClustalW | MAF
>dm3.chr2L 12166970 108 + 23011544 GUGCCCAUUCUGUUGGUUAUGUUGAUCUACUUGGGCACAUGGACGAUUUCGGCAACGCCCACAGCAGAAGUGGUUAGUAAACUUUAUAAGUACAAUAAG-----UUAAGAAUC (((((((....((.((((.....)))).)).)))))))...(((.((((((....)((.....)).))))).)))....(((((............)))-----))....... ( -26.20, z-score = -0.65, R) >apiMel3.Group10 9322293 99 - 11440700 GUCCCGAUAUUGUUAGUGAUGUUAAUUUAUCUGGGCACGUGGACGCUCAGCGCACCACCAUACGCUGAAGAG------AUAGCAGACAAUUAUGGUCUGAAGUUC-------- ((((((....((((...((((......))))..)))))).))))..((((((..........)))))).(((------....(((((.......)))))...)))-------- ( -25.20, z-score = -0.44, R) >droSim1.chr2L 11966876 108 + 22036055 GUGCCCAUUCUGUUGGUUAUGCUGAUCUACUUGGGCACAUGGACGAUUUCGGCAACGCCAACAGCAGAAGUGGUUAGUGAACUUUAUAAAUAUAAUAUG-----UUCAGAAAC (((((((....((..(.....)..)).....)))))))...(((.((((((....)((.....)).))))).)))..(((((.((((....))))...)-----))))..... ( -30.10, z-score = -1.82, R) >droSec1.super_16 360026 108 + 1878335 GUGCCCAUUCUGUUGGUUAUGCUGAUCUACUUGGGCACAUGGACGAUUUCGGCAACGCCAACAGCAGAAGUGGUUAGUAAACUUAAUAGCUAUAAUAUG-----UUAAGAAAC (((((((....((..(.....)..)).....)))))))...(((.((((((....)((.....)).))))).)))......(((((((.........))-----))))).... ( -29.70, z-score = -1.66, R) >droYak2.chr2L 8586722 113 + 22324452 GUGCCCAUCCUGUUGGUUAUGCUCAUCUACUUGGGCACAUGGACCAUUUCGGCCACGCCCACAGCAGAAGUGGUAAGUGCAUUUUAUAAUAAUUAUAUGUGAUAUUAAGGAAU .......((((....((((((((((......))))).(((..((((((((.((..........)).))))))))..)))...................)))))....)))).. ( -28.40, z-score = -0.06, R) >droEre2.scaffold_4929 13395587 113 - 26641161 GUGCCCAUUCUGUUGGUCAUGCUCAUCUACUUGGGCACAUGGACGAUUUCGGCAACGCCCACAGCAGAAGUGGUAAGUAUAUUUCCUAAAAAUUAUAUGAGAUUCUAGGGAAU .((((..(((((((((((.((((((......))))))....))).....((....))....))))))))..)))).....((((((((..((((......)))).)))))))) ( -33.70, z-score = -1.96, R) >droAna3.scaffold_12916 2966228 108 - 16180835 GUGCCCAUACUCCUGGUAAUGCUCAUCUAUUUGGGCACUUGGACUAUAUCAGCUACUCCGUAUGCGGAAGUGGUCAGUAGACUUAACGACUAUACAACUUGGUGGUUC----- (((((((......(((......)))......))))))).....((((....(((((((((....))).))))))..))))....(((.((((.......)))).))).----- ( -29.90, z-score = -0.88, R) >dp4.chr4_group3 5684758 103 - 11692001 GUGCCCAUUCUGUUGGUAAUGCUGAUCUAUUUGGGUACCUGGACCAUCUCGGCCACGCCCUAUGCGGAGGUGGUAAGUGGGAGACAGGGAAAAUUUAAGAUUU---------- .((((((....((..(.....)..)).....))))))((((..((((....(((((..((.....))..)))))..))))....))))...............---------- ( -30.70, z-score = -0.07, R) >droPer1.super_1 2781666 103 - 10282868 GUGCCCAUUCUGUUGGUAAUGCUGAUCUAUUUGGGUACCUGGACCAUCUCGGCCACGCCCUAUGCGGAGGUGGUAAGUGAGAGACAGGGAAAAUGUAAGAAAA---------- .((((((....((..(.....)..)).....))))))((((.....((((((((((..((.....))..)))))...)))))..))))...............---------- ( -29.90, z-score = -0.35, R) >droVir3.scaffold_12963 8381518 108 + 20206255 GUGCCCAUAUUGCUCGUAAUGCUCAUCUAUCUGGGCACUUGGACUAUUUCGGCCACGCCUUAUGCUGAGGUGGUAAGUAUAAGAUAUGAAGGAAGAAUGAAAGGAUUU----- .....(((.((.((((((.((((((......))))))((((.(((((((((((..........))))))))))).....)))).)))).)).))..))).........----- ( -25.00, z-score = 0.52, R) >droMoj3.scaffold_6500 993610 103 + 32352404 GUGCCCAUCUUGCUCGUAAUGCUCAUCUAUCUGGGCACCUGGACAAUUUCCGCCACGCCCUACGCUGAGGUGGUAAGU----GACGGAGAGAAGAGUUGAAUCAAUU------ (((((((....((.......)).........))))))).(.(((..((((((.(((...((((......))))...))----).)))))).....))).).......------ ( -26.92, z-score = 0.39, R) >droGri2.scaffold_15252 11584082 100 - 17193109 GUUCCCAUACUGCUUGUAAUGCUCAUCUAUCUGGGCACAUGGACUAUUUCGGCCACGCCCAUGGCGGAGGUGGUAAGUG--GCACAAGCUUAUAAUUGGAAU----------- (((((......((((((..((((((......))))))(((..((((((((.((((......)))).))))))))..)))--..))))))........)))))----------- ( -37.04, z-score = -2.62, R) >droWil1.scaffold_180708 9842337 105 + 12563649 GUGCCGAUUCUUCUGGUGAUGUUAAUCUAUUUGGGUACUUGGACCAUCUCGGCCACGCCCUAUGCAGAGGUGGUAAGUAUAGCAAGCAAAUAGGGGCUGAAAUGU-------- ..(((((....))............((((((((.((((((..((((((((.((..........)).))))))))))))))......)))))))))))........-------- ( -27.00, z-score = 0.75, R) >anoGam1.chr3R 39057531 82 - 53272125 GUCCCGAUACUGCUGGUCAUGUUUAUCUACCUUGGCACCUGGACACUGUCGGCACCGCCGACCGCAGAACGGGUAAGUUGAC------------------------------- ..((((...((((..((((.((......))..))))...........((((((...)))))).))))..)))).........------------------------------- ( -24.00, z-score = -0.17, R) >consensus GUGCCCAUUCUGUUGGUAAUGCUCAUCUAUUUGGGCACAUGGACGAUUUCGGCCACGCCCAAAGCAGAAGUGGUAAGUAAACUACAUAAAUAUAAUAUGAAAU_UU_______ (((((((........................)))))))....((.(((((.((..........)).))))).))....................................... (-13.23 = -13.60 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:30 2011