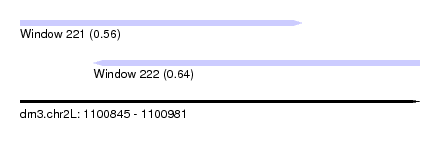
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,100,845 – 1,100,981 |
| Length | 136 |
| Max. P | 0.643502 |

| Location | 1,100,845 – 1,100,941 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.82 |
| Shannon entropy | 0.43591 |
| G+C content | 0.49595 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -15.31 |
| Energy contribution | -14.61 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
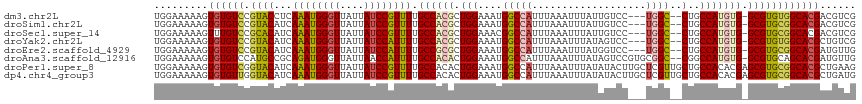
>dm3.chr2L 1100845 96 + 23011544 ACGAUGAUUUAGGCACUUCAUCCUC-------CGACGUC--------GUGCCACACGC-CACAUGGCAG--GCCA---GGACAAUAAAUUUAAAUGGCCAUUUCCAGCGUGGCAAAA ((((((....(((........))).-------...))))--------))(((((.(((-(....))).(--((((---................))))).......).))))).... ( -30.69, z-score = -2.52, R) >droSim1.chr2L 1071296 96 + 22036055 ACGAUGAUUUAGGCACCUCAUCCUC-------CGACGUC--------GUGCCGCACGC-CACAUGGCAG--GCCA---GGACAAUAAAUUUAAAUGGCCAUUUCCAGCGUGGCAAAA ((((((....(((........))).-------...))))--------))(((((.(((-(....))).(--((((---................))))).......).))))).... ( -30.29, z-score = -1.74, R) >droSec1.super_14 1054303 96 + 2068291 ACGAUGAUUUAGGCACCUCAUCCUC-------CGACGUC--------GUGCCGCACGC-CACAUGGCAG--GCCA---GGACAAUAAAUUUAAAUGGCCGUUUCCAGCGUGGCAAAA ...........(((((..(.((...-------.)).)..--------)))))....((-(((.(((..(--((((---................)))))....)))..))))).... ( -30.89, z-score = -1.58, R) >droYak2.chr2L 1078007 96 + 22324452 ACUUCAUCAUGGGCACUUCAUCCUC-------CGACAGC--------GUGCCACACGC-CACAUGGCAG--GCCA---GGACUAUAAAUUUAAAUGGCCAUUUCCAGCGUGGCAAAA .........(((((((....((...-------.))....--------))))).)).((-(((.(((..(--((((---................)))))....)))..))))).... ( -27.79, z-score = -1.46, R) >droEre2.scaffold_4929 1145119 96 + 26641161 ACGAUGAUUUGGGUACUUUAUCCUG-------CAACAUC--------GUGCCGCACGC-CACAUGGCAG--GCCA---GGACCAUAAAUUUAAAUGGCCAUUUCCAGCGCGGCAAAA ((((((....(((((...)))))..-------...))))--------))(((((.(((-(....))).(--((((---................))))).......).))))).... ( -34.39, z-score = -2.79, R) >droAna3.scaffold_12916 4567444 106 + 16180835 AUGAUGACUUAACCACUCCAUCCUUAGCCUUGCAACAUC--------GUGCUGCACGC-CACAUGGCCG--GCCGCACGGACUAUAAAUUUAAAUGGCCAUUUCCAGUGUGGCAAAA ..((((............))))....(((.(((......--------((((.((..((-(....)))..--)).))))((.((((........)))))).......))).))).... ( -26.30, z-score = -0.33, R) >droPer1.super_8 732299 117 + 3966273 AUCAUGACUUAACCAUUACAUACUGAAGCUUGCCACAGCACUUCAGCGUGCCGCACGCUCGUGUGGCAGCAACGAGCAAGUAUAUAAAUUUAAAUGGCCAUUUCCAGUGUGGCAAAA ......................((((((..(((....)))))))))..((((((((((((((((....)).)))))).................(((......)))))))))))... ( -34.50, z-score = -1.72, R) >dp4.chr4_group3 9544883 117 + 11692001 AUCAUGACUUAGCCAUUACAUACUGAAGCUUGCCACAGCACAUCAGCGUGCCGCACGCUCGUGUGGCAGCAACGAGCAAGUAUAUAAAUUUAAAUGGCCAUUUCCAGUGUGGCAAAA ...........(((((.(((((((...((((((....))......((.((((((((....))))))))))...)))).)))))...........(((......)))))))))).... ( -35.00, z-score = -1.40, R) >consensus ACGAUGACUUAGGCACUUCAUCCUC_______CGACAUC________GUGCCGCACGC_CACAUGGCAG__GCCA___GGACUAUAAAUUUAAAUGGCCAUUUCCAGCGUGGCAAAA ................................................((((((.(......(((((.............................))))).....).))))))... (-15.31 = -14.61 + -0.70)



| Location | 1,100,870 – 1,100,981 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.88 |
| Shannon entropy | 0.25437 |
| G+C content | 0.47538 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -24.01 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1100870 111 - 23011544 UGGAAAAAGUGUGUCCGUACCUCAAAUGGGUUAUUAUCCGUUUUGCCACGCUGGAAAUGGCCAUUUAAAUUUAUUGUCC---UGGC--CUGCCAUGUG-GCGUGUGGCACGACGUCG ..((.....((((.((((((...((((((((....)))))))).((((((.(((....(((((..(((.....)))...---))))--)..)))))))-))))))))))))...)). ( -37.50, z-score = -2.09, R) >droSim1.chr2L 1071321 111 - 22036055 UGGAAAAAGUGUGUCCGUACAUCAAAUGGGUUAUUAUCCGUUUUGCCACGCUGGAAAUGGCCAUUUAAAUUUAUUGUCC---UGGC--CUGCCAUGUG-GCGUGCGGCACGACGUCG ..((.....((((.((((((...((((((((....)))))))).((((((.(((....(((((..(((.....)))...---))))--)..)))))))-))))))))))))...)). ( -43.20, z-score = -3.44, R) >droSec1.super_14 1054328 111 - 2068291 UGGAAAAAGUUUGUCCGCACAUCAAAUGGGUUAUUAUCCGUUUUGCCACGCUGGAAACGGCCAUUUAAAUUUAUUGUCC---UGGC--CUGCCAUGUG-GCGUGCGGCACGACGUCG ..((....(((((.((((((...((((((((....)))))))).((((((.(((....(((((..(((.....)))...---))))--)..)))))))-)))))))))).))).)). ( -41.90, z-score = -2.88, R) >droYak2.chr2L 1078032 111 - 22324452 UGGAAAAAGUGUGUCCGUACAUCAAAUGGGUUAUUAUCCAUUUUGCCACGCUGGAAAUGGCCAUUUAAAUUUAUAGUCC---UGGC--CUGCCAUGUG-GCGUGUGGCACGCUGUCG ..((...((((((.((((((...((((((((....)))))))).((((((.(((....(((((.(((......)))...---))))--)..)))))))-)))))))))))))).)). ( -43.80, z-score = -4.07, R) >droEre2.scaffold_4929 1145144 111 - 26641161 UGGAAAAAGUGUGUCCGUACAUCAAAUGGGUUAUUAUCCAUUUUGCCGCGCUGGAAAUGGCCAUUUAAAUUUAUGGUCC---UGGC--CUGCCAUGUG-GCGUGCGGCACGAUGUUG .(((.........)))..(((((((((((((....))))))))((((((((.((..(.((((((........)))))).---)..)--).(((....)-)))))))))).))))).. ( -42.20, z-score = -2.89, R) >droAna3.scaffold_12916 4567476 114 - 16180835 UGGAAAAAGUGUGUCCAUGCCGCAGAUGGGUUAUUAACCAUUUUGCCACACUGGAAAUGGCCAUUUAAAUUUAUAGUCCGUGCGGC--CGGCCAUGUG-GCGUGCAGCACGAUGUUG ........((((...((((((((((((((........))).))))).(((.(((...(((((.....................)))--)).)))))))-)))))..))))....... ( -35.40, z-score = -0.18, R) >droPer1.super_8 732339 117 - 3966273 UGGAAAAAGUGUGUCGGUACAUCAAAUGGGUUAUUAUCCGUUUUGCCACACUGGAAAUGGCCAUUUAAAUUUAUAUACUUGCUCGUUGCUGCCACACGAGCGUGCGGCACGCUGAAG .......((((((((((((....((((((((....)))))))))))).((((((......))).................((((((((....)).))))))))).)))))))).... ( -34.50, z-score = -1.23, R) >dp4.chr4_group3 9544923 117 - 11692001 UGGAAAAAGUGUGUUGGUACAUCAAAUGGGUUAUUAUCCGUUUUGCCACACUGGAAAUGGCCAUUUAAAUUUAUAUACUUGCUCGUUGCUGCCACACGAGCGUGCGGCACGCUGAUG ..((..(((((((((((((....((((((((....)))))))))))))...(((......))).........))))))))..))...((((((.(((....))).)))).))..... ( -32.50, z-score = -0.55, R) >consensus UGGAAAAAGUGUGUCCGUACAUCAAAUGGGUUAUUAUCCGUUUUGCCACGCUGGAAAUGGCCAUUUAAAUUUAUAGUCC___UGGC__CUGCCAUGUG_GCGUGCGGCACGAUGUCG .........((((((.((((...((((((((....)))))))).((((((.(((.....((((...................)))).....))))))).))))))))))))...... (-24.01 = -24.48 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:43 2011