
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,473,190 – 20,473,281 |
| Length | 91 |
| Max. P | 0.529951 |

| Location | 20,473,190 – 20,473,281 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 58.98 |
| Shannon entropy | 0.79261 |
| G+C content | 0.45804 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -5.46 |
| Energy contribution | -6.21 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
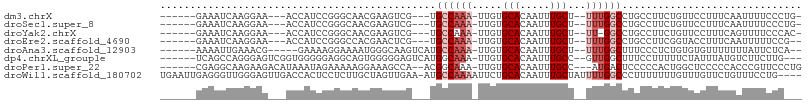
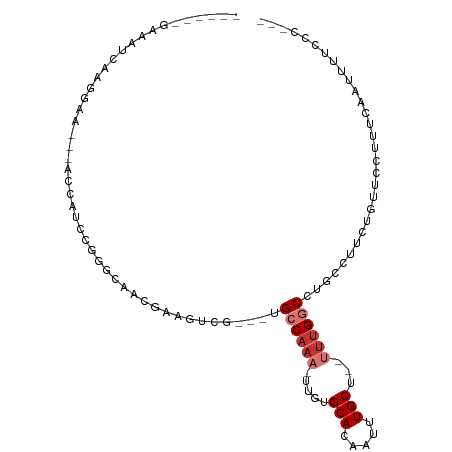
>dm3.chrX 20473190 91 + 22422827 ------GAAAUCAAGGAA---ACCAUCCGGGCAACGAAGUCG---UGCCAAA-UUGUGCACAAUUUGCU--UUUGGCCUGCCUUCUGUUCCUUUCAAUUUUCCCUG- ------.....((.((((---(......(((.(((((((.((---.((((((-....(((.....))).--)))))).)).)))).))))))......))))).))- ( -23.60, z-score = -1.93, R) >droSec1.super_8 2714051 91 + 3762037 ------GAAAUCAAGGAA---ACCAUCCGGGCAACGAAGUCG---UGCCAAA-UUGUGCACAAUUUGCU--UUUGGCCUGCCUUCUGUUCCUUUCAAUUUUCCCUG- ------.....((.((((---(......(((.(((((((.((---.((((((-....(((.....))).--)))))).)).)))).))))))......))))).))- ( -23.60, z-score = -1.93, R) >droYak2.chrX 19702240 90 + 21770863 ------GAAAUCAAGGAA---ACCAUCCGGGCAACGAAGUCG---UGCCAAA-UUGUGCACAAUUUGCU--UU-GGCCUGCCUUCUGUUCCUUUCAGUUUUCCCAC- ------........((((---(.(....(((.(((((((.((---.((((((-....(((.....))))--))-))).)).)))).))))))....).)))))...- ( -22.00, z-score = -0.94, R) >droEre2.scaffold_4690 17074393 90 + 18748788 ------GAAAUCAAGGAA---ACCAUCCGGGCCACGAACUCG---UGCCAAA-UUGUGCACAAUUUGCU--UUUGGCCUGCCUUCGGUACCUUUCAAUUUUUCCG-- ------........((((---(.....(((((((((....))---)((.(((-(((....)))))))).--...)))))).(....)............))))).-- ( -23.10, z-score = -1.63, R) >droAna3.scaffold_12903 551520 91 - 802071 ------AAAAUUGAAACG-----GAAAAGGAAAAUGGGCAAGUCAUGCCAAA-UUGUGCACAAUUUGCU--UUUGGCUUUCCCUCUGUGUGUUUUUUUAUUCUCA-- ------......((((((-----.(...(((....(((.((((((.((.(((-(((....)))))))).--..)))))).)))))).).))))))..........-- ( -21.90, z-score = -1.13, R) >dp4.chrXL_group1e 3417493 95 - 12523060 ------UCAGCCAGGGAGUCGGUGGGGGAGGCAGUGGGGGAGUCAUGGCAAA-UUGUGCACAAUUUGCC--GUUGGCUUUCCUUUUUCUAUUUAUGUCUUCUUG--- ------.....(((((((.((.....(((((....(((..((((((((((((-(((....)))))))))--).)))))..))).))))).....)).)))))))--- ( -39.10, z-score = -3.96, R) >droPer1.super_22 585591 95 - 1688296 ------CGAGGCAAGAAGACAUAAAUAGAAAAAGGAAAGCCA--ACGGCAAA-UUGUGCACAAUUUGCC---AUGACUCCCCCACUGGCUCCCCCACCCGUUCCCUG ------...........................((..(((((--..((((((-(((....)))))))))---.((.......)).)))))..))............. ( -20.60, z-score = -1.10, R) >droWil1.scaffold_180702 2818409 102 + 4511350 UGAAUUGAGGGUUGGGAGUUGACCACUCCUCUUGCUAGUUGAA-AUGCCAAAAUUCUGCACAAUUUGCUAUUUUGGCCCUUUUUUUGUUUGUUCUGUUUCCUG---- .(((..((((((.((((((.....))))))...))........-..((((((((...(((.....))).))))))))))))..))).................---- ( -23.90, z-score = -1.10, R) >consensus ______GAAAUCAAGGAA___ACCAUCCGGGCAACGAAGUCG___UGCCAAA_UUGUGCACAAUUUGCU__UUUGGCCUGCCUUCUGUUCCUUUCAAUUUUCCC___ ..............................................((((((.....(((.....)))...)))))).............................. ( -5.46 = -6.21 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:14 2011