| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,454,033 – 20,454,187 |
| Length | 154 |
| Max. P | 0.973922 |

| Location | 20,454,033 – 20,454,187 |
|---|---|
| Length | 154 |
| Sequences | 5 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 68.94 |
| Shannon entropy | 0.54896 |
| G+C content | 0.51496 |
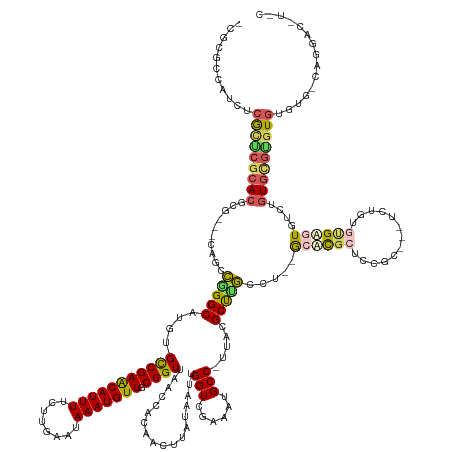
| Mean single sequence MFE | -53.60 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
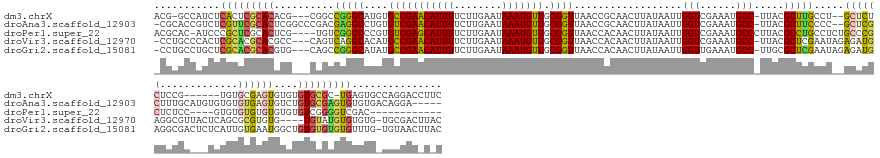
>dm3.chrX 20454033 154 + 22422827 ACG-GCCAUCUCACUCGCACACG---CGGCCGGGCAUGUGCCGAACAUUUUCUUGAAUAAAUGUUGCGGUUAACCGCAACUUAUAAUUGGUCGAAAUGCC-UUACGCUUGCCU--GCUCUCUCCG------UGUGCGAGUGUGUGUGCGC-UGAGUGCCAGGACCUUC ..(-(((..(.((((((((((((---.((.((((((.(((.....((((((..(.(((....(((((((....))))))).....))).)..))))))..-...))).)))))--)....)).))------)))))))))).)((.((((-...)))))))).))... ( -60.30, z-score = -3.43, R) >droAna3.scaffold_12903 532425 159 - 802071 -CGCACCGUCUCGUUCGCACUCGGCCCGACGAGGCCUGUGUCGAACAUUUUCUUGAAUAAAUGUUGCGGUUAACCGCAACUUAUAAUUGGUCGAAAUGCC-UUACGCUUCCCC--GCUCGCUUUGCAUGUGUGUGUGAGUGUCUGUGCGAGUGUGUGACAGGA----- -(((((...(((((((((((..((((......)))).))).)))))(((((..(.(((....(((((((....))))))).....))).)..)))))(((-..((((((...(--((.(((.......))).))).))))))..).))))).)))))......----- ( -53.60, z-score = -1.66, R) >droPer1.super_22 564688 147 - 1688296 ACGCAC-AUCCCGCUCGCACUCG----UGUCGGGCCCGUGUCGAGCAUUUUCUUGAAUAAAUGUUGCGGUUAACCACAACUUAUAAUUGGUCGAAAUGCCCUUACGCCUGCCUCUGCCCGCUCUCC----GUGUGUGUGUGUGUGUCGGGGUCGAC------------ .((...-(((((((.(((((.((----.(.(((((..(((..(.(((((((..(.(((....((((.((....)).)))).....))).)..))))))))..)))))))).).).((.(((.....----))).))).))))).).))))))))..------------ ( -42.90, z-score = 0.03, R) >droVir3.scaffold_12970 4742491 158 + 11907090 -CCUGCCCACUCGCACGCACGCC---CAGUCAGGCACAUGCCGAACAUUUUCUUGAAUAAAUGUUGCGGUUAACCACAACUUAUAAUUGGUCGAAAUGCC-UUACGCUCGAAUAGAGAUGAGGCGUUACUCAGCGCGUGUG----UGUAUGUGUGUG-UGCGACUUAC -.........((((((((((((.---..((.(.((((((((..(((((((........)))))))((..(..((((...........))))..)((((((-(((..(((.....))).))))))))).....)))))))))----).))))))))))-)))))..... ( -54.40, z-score = -2.63, R) >droGri2.scaffold_15081 3698587 162 - 4274704 -CCUGCCUGCUCGCACGCACGUG---CAGCCGGGCAUAUGCCGAACAUUUUCUUGAAUAAAUGUUGCGGUUAACCACAACUUAUAAUUGGUUGAAAUGCC-UUGCGCUCGAAUAGAGAUGAGGCGACUCUCAUUGUGAAUGGCUGUGUGUGUGUUUG-UGUAACUUAC -......(((.((.(((((((..---((((((..((((.(((((((((((........))))))).))))......(((((.......)))))...((((-((...(((.....)))..))))))........))))..))))))..))))))).))-.)))...... ( -56.80, z-score = -3.35, R) >consensus _CGCGCCAUCUCGCUCGCACGCG___CAGCCGGGCAUGUGCCGAACAUUUUCUUGAAUAAAUGUUGCGGUUAACCACAACUUAUAAUUGGUCGAAAUGCC_UUACGCUUGCCU__GCACGCUCCGC___UCUGUGUGAGUGUCUGUGCGUGUGUGUG_CAGGAC_U_C ...........(((((((((..........(((((.....((((((((((........))))))).)))(..((((...........))))..)...........))))).....((((((.............))))))....)))))))))............... (-24.38 = -24.46 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:13 2011