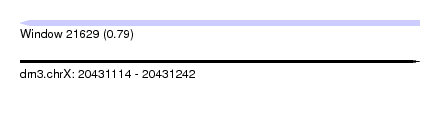
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,431,114 – 20,431,242 |
| Length | 128 |
| Max. P | 0.792156 |

| Location | 20,431,114 – 20,431,242 |
|---|---|
| Length | 128 |
| Sequences | 7 |
| Columns | 147 |
| Reading direction | reverse |
| Mean pairwise identity | 55.00 |
| Shannon entropy | 0.84545 |
| G+C content | 0.55494 |
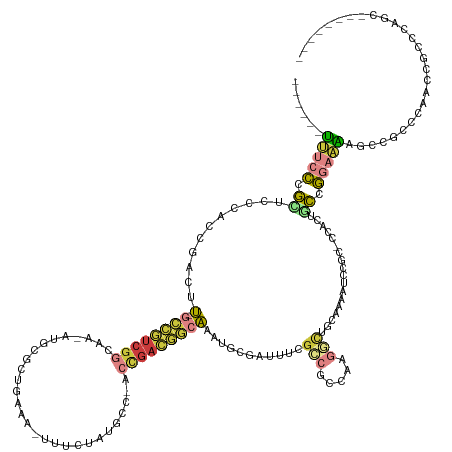
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -10.83 |
| Energy contribution | -11.59 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.72 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
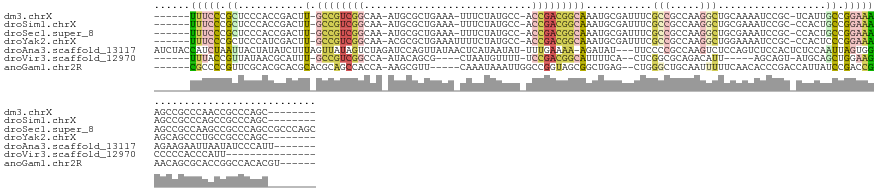
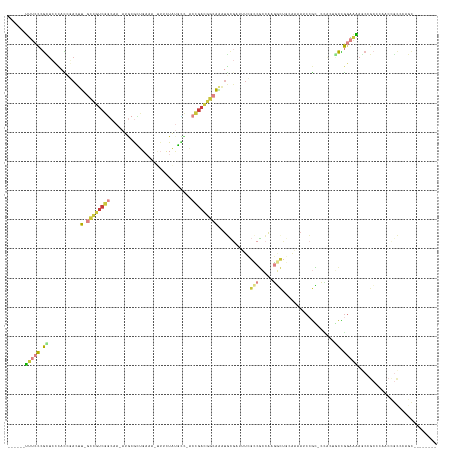
>dm3.chrX 20431114 128 - 22422827 ------UUUCCCGCUCCCACCGACUU-GCCGUCGGCAA-AUGCGCUGAAA-UUUCUAUGCC-ACCGACGGCAAAUGCGAUUUCGCCGCCAAGGCUGCAAAAUCCGC-UCAUUGCCGGAAAAGCCGCCCAACCGCCCAGC-------- ------(((((.((..........((-((((((((...-..(((..((..-..))..))).-.))))))))))..(((((((.((.((....)).)).)))).)))-.....)).)))))...................-------- ( -38.60, z-score = -1.24, R) >droSim1.chrX 15659808 128 - 17042790 ------UUUCCCGCUCCCACCGACUU-GCCGUCGGCAA-AUGCGCUGAAA-UUUCUAUGCC-ACCGACGGCAAAUGCGAUUUCGCCGCCAAGGCUGCGAAAUCCGC-CCACUGCCGGAAAAGCCGCCCAGCCGCCCAGC-------- ------(((((.((..........((-((((((((...-..(((..((..-..))..))).-.))))))))))..((((((((((.((....)).))))))).)))-.....)).))))).((.((...)).)).....-------- ( -45.80, z-score = -2.74, R) >droSec1.super_8 2662022 136 - 3762037 ------UUUCCCGCUCCCACCGACUU-GCCGUCGGCAA-AUGCGCUGAAA-UUUCUAUGCC-ACCGACGGCAAAUGCGAUUUCGCCGCCAAGGCUGCGAAAUCCGC-CCACUGCCGGAAAAGCCGCCAAGCCGCCCAGCCGCCCAGC ------(((((.((..........((-((((((((...-..(((..((..-..))..))).-.))))))))))..((((((((((.((....)).))))))).)))-.....)).)))))....((...((.((...)).))...)) ( -46.50, z-score = -1.94, R) >droYak2.chrX 19658147 129 - 21770863 ------UUUCCCGCUCCCAUCGACUU-GCCGUCGGCAA-ACGCGCUGAAAUUUUCUAUGCC-ACCGACGGCAAAUGCGAUUUCGCCGCCAAGGCUGGAAAAUCCGC-CCACUCCCGGAAAAGCAGCCCUGCCGCCCAGC-------- ------......(((...((((..((-((((((((...-..(((..((.....))..))).-.))))))))))...))))...((.((...(((((.....((((.-.......))))....)))))..)).))..)))-------- ( -40.40, z-score = -1.35, R) >droAna3.scaffold_13117 1533631 135 + 5790199 AUCUACCAUCUAAUUACUAUAUCUUUAGUUAUAGUCUAGAUCCAGUUAUAACUCAUAAUAU-UUUGAAAA-AGAUAU---UUCCCCGCCAAGUCUCCAGUCUCCACUCUCCAAUUAGUGGAGAAGAAUUAAUAUCCCAUU------- ..........(((((.((((((((((.(((((((.((......)))))))))(((......-..))).))-))))))---...................((((((((........)))))))))))))))..........------- ( -23.10, z-score = -2.68, R) >droVir3.scaffold_12970 4717135 111 - 11907090 ------UUUACCGUUAUAACGCAUUU-GCCGUCGGCCA-AUACAGCG----CUAAUGUUUU-UCCGACGGCAUUUUCA--CUCGGCGCAGACAUU-----AGCAGU-AUGCAGCUGGAAGCCCCCACCCAUU--------------- ------..............((...(-((((((((...-..(((...----....)))...-.)))))))))......--.((((((((.((...-----....))-.))).)))))..))...........--------------- ( -29.20, z-score = -1.25, R) >anoGam1.chr2R 8234452 127 - 62725911 ------CGCCCCGUUCGCACGCACGCACGCAGCCACCA-AAGCGUU-----CAAAUAAAUUGGCCGGUAGCGGCUGAG--CUGGGCUGCAAUUUUUCAACACCCGACCAUUAUCCGACCGAACAGCGCACCGGCCACACGU------ ------.((..(((.((......)).)))..)).....-.......-----.........((((((((.(((.(((..--.((((.((...........)))))).......((.....)).)))))))))))))).....------ ( -31.90, z-score = 1.72, R) >consensus ______UUUCCCGCUCCCACCGACUU_GCCGUCGGCAA_AUGCGCUGAAA_UUUCUAUGCC_ACCGACGGCAAAUGCGAUUUCGCCGCCAAGGCUGCAAAAUCCGC_CCACUGCCGGAAAAGCCGCCCAACCGCCCAGC________ ......(((((.((.............((((((((............................))))))))............(((.....)))..................)).)))))........................... (-10.83 = -11.59 + 0.76)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:09 2011