| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,422,212 – 20,422,302 |
| Length | 90 |
| Max. P | 0.599618 |

| Location | 20,422,212 – 20,422,302 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.86 |
| Shannon entropy | 0.34077 |
| G+C content | 0.42950 |
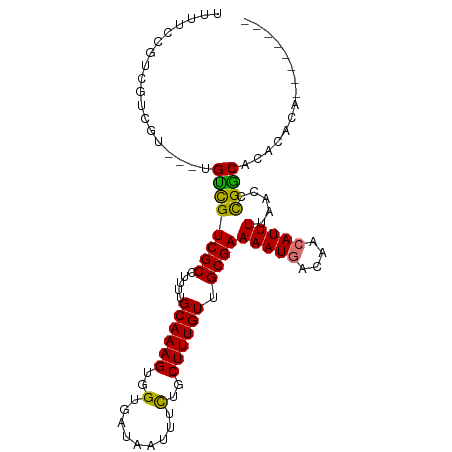
| Mean single sequence MFE | -22.01 |
| Consensus MFE | -16.18 |
| Energy contribution | -15.72 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
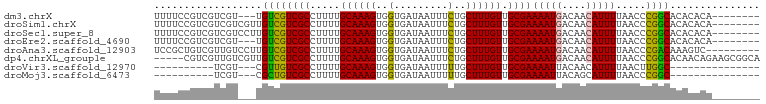
>dm3.chrX 20422212 90 + 22422827 UUUUCCGUCGUCGU---UGUCGUCGCCUUUUGCAAAGUGGUGAUAAUUUCUGCUUUGUUGCGAAAAUGACAACAUUUUAACCCGGCACACACA-------- ......((((..((---(((((((((.....((((((..(.........)..)))))).))))...))))))).........)))).......-------- ( -22.00, z-score = -1.05, R) >droSim1.chrX 15650015 93 + 17042790 UUUUCCGUCGUCGUCGUUGUCGUCGCCUUUUGCAAAGUGGUGAUAAUUUCUGCUUUGUUGCGAAAAUGACAACAUUUUAACCCGGCACACACA-------- ......((((.....(((((((((((.....((((((..(.........)..)))))).))))...))))))).........)))).......-------- ( -22.74, z-score = -0.88, R) >droSec1.super_8 2617731 93 + 3762037 UUUUCCGUCGUCGUCCUUGUCGUCGCCUUUUGCAAAGUGGUGAUAAUUUCUGCUUUGUUGCGAAAAUGACAACAUUUUAACCCGGCACACACA-------- ......((((.......(((.((((..((((((((...(((((.....)).)))...)))))))).)))).)))........)))).......-------- ( -20.36, z-score = -0.59, R) >droEre2.scaffold_4690 17022983 90 + 18748788 UUUUCCGUCGUCGU---UGUCGUCGCCUUUUGCAAAGUGGUGAUAAUUUCUGCUUUGUUGCGAAAAUGACAACAUUUUAACCCGGCACACACA-------- ......((((..((---(((((((((.....((((((..(.........)..)))))).))))...))))))).........)))).......-------- ( -22.00, z-score = -1.05, R) >droAna3.scaffold_12903 491127 92 - 802071 UCCGCUGUCGUUGUCCUUGUCGUCGCCUUUUGCAAAGUGGUGAUAAUUUCUGCUUUGUUGCGAAAAUGACAACAUUUUAACCCGACAAAGUC--------- .....((((((((((.((....((((.....((((((..(.........)..)))))).)))).)).))))))..........)))).....--------- ( -21.50, z-score = -1.61, R) >dp4.chrXL_group1e 9048802 96 + 12523060 -----CGUCGUUGUCGUUGUCGUCGCCUUUUGCAAAGUGGUGAUAAUUUCUGCUUUGUUGCGAAAAUGACAACAUUUUAACCCGGCACAACAGAAGCGGCA -----.((((((((((((....((((.....((((((..(.........)..)))))).)))).)))))))))...........((.........))))). ( -28.80, z-score = -1.33, R) >droVir3.scaffold_12970 4706631 73 + 11907090 ----------UCGU---CGUUGUCGCCUUUUGCAAAGUGGUGAUAAUUUUUGCUUUGUUGCGAAAAUUACAACAUUUUAACUUGGC--------------- ----------....---.(((((((((...........)))))((((((((((......)))))))))))))).............--------------- ( -18.00, z-score = -1.86, R) >droMoj3.scaffold_6473 4137497 73 + 16943266 ----------UCGU---CGCUGUCGCCUUUUGCAAAGUGGUGAUAAUUUUUGCUUUGUUGCGAAAAUUACAGCAUUUUAACCCGGC--------------- ----------....---.((((..((.....))((((((.((.((((((((((......)))))))))))).))))))....))))--------------- ( -20.70, z-score = -2.09, R) >consensus UUUUCCGUCGUCGU___UGUCGUCGCCUUUUGCAAAGUGGUGAUAAUUUCUGCUUUGUUGCGAAAAUGACAACAUUUUAACCCGGCACACACA________ ..................((((((((.....((((((..(.........)..)))))).))))(((((....))))).....))))............... (-16.18 = -15.72 + -0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:08 2011