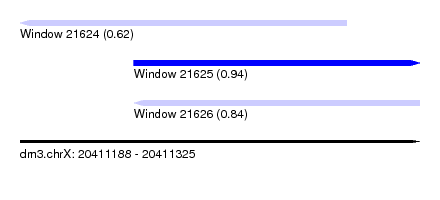
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,411,188 – 20,411,325 |
| Length | 137 |
| Max. P | 0.938526 |

| Location | 20,411,188 – 20,411,300 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 74.53 |
| Shannon entropy | 0.47840 |
| G+C content | 0.43456 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -19.49 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
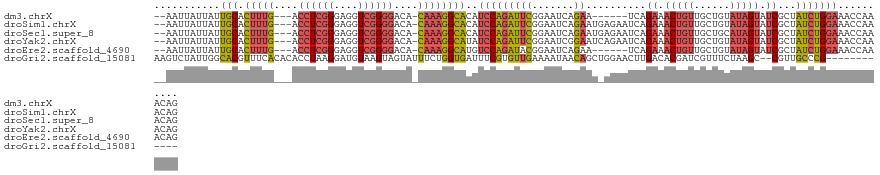
>dm3.chrX 20411188 112 - 22422827 --AAUUAUUAUUGCACUUUG---ACCUCGGGAGGUCGGGGACA-CAAAGGCACAUCCAGAUUCGGAAUCAGAA------UCAGAAACUGUUGCUGUAUAGUAUCGCUAUCUGGAAACCAAACAG --.........(((.(((((---((((....)))))(....).-.)))))))..(((((((.(((.(((((..------.(((...)))...)))....)).)))..))))))).......... ( -28.00, z-score = -1.23, R) >droSim1.chrX 15638713 118 - 17042790 --AAUUAUUAUUGCACUUUG---ACCUCGGGAGGUCGGGGACA-CAAAGGCACAUCCAGAUUCGGAAUCAGAAUGAGAAUCAGAAACUGUUGCUGUAUAGUAUCGCUAUCUGGAAACCAAACAG --.........(((.(((((---((((....)))))(....).-.)))))))..(((((((.(.((.((.......)).)).((.(((((......))))).)))..))))))).......... ( -28.50, z-score = -0.81, R) >droSec1.super_8 2607164 118 - 3762037 --AAUUAUUAUUGCACUUUG---ACCUCGGGAGGUCGGGGACA-CAAAGGCACAUCCAGAUUCGGAAUCAGAAUGAGAAUCAGAAACUGUUGCUGCAUAGUAUCGCUAUCUGGAAACCAAACAG --.........(((.(((((---((((....)))))(....).-.)))))))..(((((((.(.((.((.......)).)).((.(((((......))))).)))..))))))).......... ( -27.80, z-score = -0.58, R) >droYak2.chrX 19635883 118 - 21770863 --AAUUAUUAUUGCACUUUG---ACCUCGGGAGGUCGGGGACA-CAAAGGCAUAUCCAGAUUCGGAAUCGGAAUCAGAAUCAGAAACUGUUGCUGUAUAGUAUCGCUAUCUGGAAACCAAACAG --.........(((.(((((---((((....)))))(....).-.)))))))..(((((((.(.((.((.......)).)).((.(((((......))))).)))..))))))).......... ( -29.30, z-score = -1.09, R) >droEre2.scaffold_4690 17012581 112 - 18748788 --AAUUAUUAUUGCACUUUG---ACCUCGGGAGGUCGGGGACA-CAAAGGCAUGUCCAGAUACGGAAUCAGAA------UCAGAAACUGUUGCUGUAUAGUAUCGCUAUCUGGAAACCAAACAG --.........(((.(((((---((((....)))))(....).-.)))))))..(((((((((((.(((((..------.(((...)))...)))....)).))).)))))))).......... ( -29.80, z-score = -1.57, R) >droGri2.scaffold_15081 3654606 110 + 4274704 AAGUCUAUUGGCACGUUUCACACACCUAAGGAUGUAAGUAGUAUUUCUGGUGAUUUCGUGUUGAAAAUAACAGCUGGAACUUGACACGAUCGUUUCUAAGC--CGUUGCCCG------------ .......(..(((((..(((((((((...)).)))...(((.....)))))))...)))))..)...((((.(((((((..(((.....))).)))).)))--.))))....------------ ( -22.80, z-score = 0.59, R) >consensus __AAUUAUUAUUGCACUUUG___ACCUCGGGAGGUCGGGGACA_CAAAGGCACAUCCAGAUUCGGAAUCAGAAU_AGAAUCAGAAACUGUUGCUGUAUAGUAUCGCUAUCUGGAAACCAAACAG ...........(((.(((((....((((((....))))))....))))))))..(((((((((.......))..........((.(((((......))))).))...))))))).......... (-19.49 = -20.22 + 0.73)
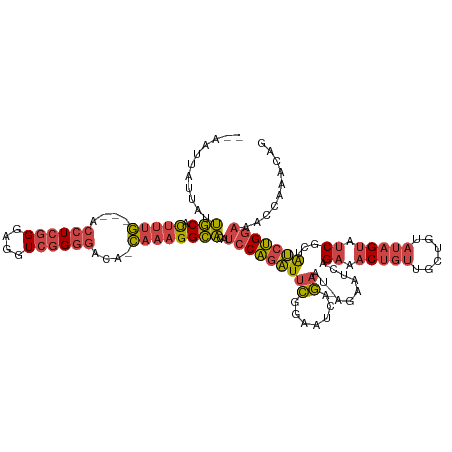
| Location | 20,411,227 – 20,411,325 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 67.67 |
| Shannon entropy | 0.63952 |
| G+C content | 0.45108 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -12.64 |
| Energy contribution | -11.70 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.81 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20411227 98 + 22422827 ------UUCUGAUUCUGAUUCCGAAUCUGGAUGUGCCUUUGUGUCCCCGACCUCCCGAGGUCAAAGUGCAAUAAUAAUUGAGCUGUUG--UCGCUGACGGCAAUAA ------....(((((.......))))).(((..(......)..)))..(((((....)))))..(((.((((....)))).)))((((--(((....))))))).. ( -24.70, z-score = -1.52, R) >droSim1.chrX 15638752 104 + 17042790 UUCUGAUUCUCAUUCUGAUUCCGAAUCUGGAUGUGCCUUUGUGUCCCCGACCUCCCGAGGUCAAAGUGCAAUAAUAAUUGAGCUGUUG--UCGCUGGCGGCAAUAA ........((((.......((((....))))..((((((((...((.((......)).)).))))).)))........)))).(((((--(((....)))))))). ( -25.80, z-score = -0.92, R) >droSec1.super_8 2607203 104 + 3762037 UUCUGAUUCUCAUUCUGAUUCCGAAUCUGGAUGUGCCUUUGUGUCCCCGACCUCCCGAGGUCAAAGUGCAAUAAUAAUUGAGCUGUUG--UCGCUGGCGGCAAUAA ........((((.......((((....))))..((((((((...((.((......)).)).))))).)))........)))).(((((--(((....)))))))). ( -25.80, z-score = -0.92, R) >droYak2.chrX 19635922 104 + 21770863 UUCUGAUUCUGAUUCCGAUUCCGAAUCUGGAUAUGCCUUUGUGUCCCCGACCUCCCGAGGUCAAAGUGCAAUAAUAAUUGAGCUGUUG--UCGCUGACGGCAAUAA ....(((((.((.......)).))))).((((((......))))))..(((((....)))))..(((.((((....)))).)))((((--(((....))))))).. ( -28.10, z-score = -2.28, R) >droEre2.scaffold_4690 17012620 98 + 18748788 ------UUCUGAUUCUGAUUCCGUAUCUGGACAUGCCUUUGUGUCCCCGACCUCCCGAGGUCAAAGUGCAAUAAUAAUUGAGCUGUUG--UCGCUGAUGGCAAUAA ------...........(((((((....((((((......))))))..(((((....)))))..(((((((((..........)))))--.)))).)))).))).. ( -23.50, z-score = -1.14, R) >droAna3.scaffold_12903 476977 77 - 802071 --------------------CCCGGCCUGGGUGU-CCUUUGUGCCCCAGGU--------ACUAAAGUGCAAUAAUAAUUGAGCUCUUGCCUCGUUGACAGCCAUAA --------------------...((((((((.((-.......)))))))((--------((....))))..........((((....).))).......))).... ( -18.90, z-score = -0.23, R) >droGri2.scaffold_15081 3654644 91 - 4274704 ------------UGUUAUUUUCAACACGAAAUCACCAGAAAUACUACUUACAUCCUUAGGUGUGUGAAACGUGCCAAUAGACUUUAUUA-AAGUUCA-GAAAAUA- ------------...(((((((..((((...((((.((.....))....(((((....)))))))))..))))......(((((.....-)))))..-)))))))- ( -14.20, z-score = -0.65, R) >consensus ______UUCUGAUUCUGAUUCCGAAUCUGGAUGUGCCUUUGUGUCCCCGACCUCCCGAGGUCAAAGUGCAAUAAUAAUUGAGCUGUUG__UCGCUGACGGCAAUAA ............................(((((........)))))..(((((....)))))...................((((((........))))))..... (-12.64 = -11.70 + -0.94)
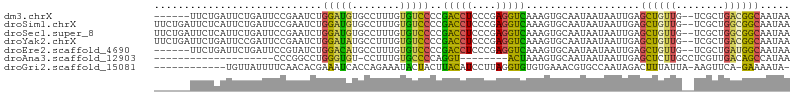
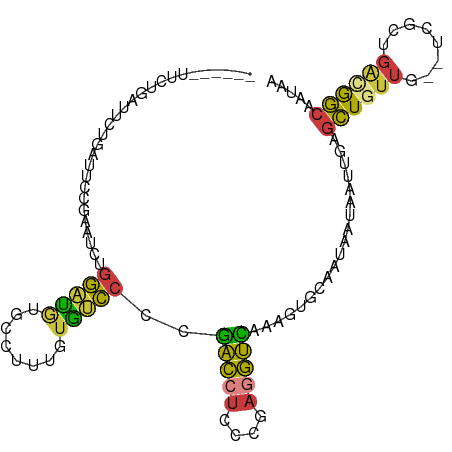
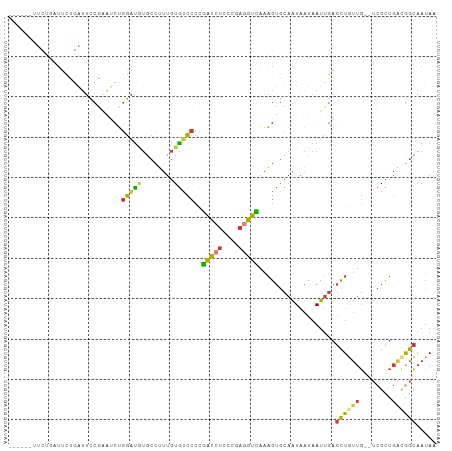
| Location | 20,411,227 – 20,411,325 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 67.67 |
| Shannon entropy | 0.63952 |
| G+C content | 0.45108 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -13.12 |
| Energy contribution | -12.63 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.61 |
| Mean z-score | -0.62 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.837555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20411227 98 - 22422827 UUAUUGCCGUCAGCGA--CAACAGCUCAAUUAUUAUUGCACUUUGACCUCGGGAGGUCGGGGACACAAAGGCACAUCCAGAUUCGGAAUCAGAAUCAGAA------ ....((((...(((..--.....)))..............(((((((((....))))))))).......))))......(((((.......)))))....------ ( -24.70, z-score = -1.51, R) >droSim1.chrX 15638752 104 - 17042790 UUAUUGCCGCCAGCGA--CAACAGCUCAAUUAUUAUUGCACUUUGACCUCGGGAGGUCGGGGACACAAAGGCACAUCCAGAUUCGGAAUCAGAAUGAGAAUCAGAA ....((((...(((..--.....)))..............(((((((((....))))))))).......))))......(((((.............))))).... ( -23.82, z-score = -0.49, R) >droSec1.super_8 2607203 104 - 3762037 UUAUUGCCGCCAGCGA--CAACAGCUCAAUUAUUAUUGCACUUUGACCUCGGGAGGUCGGGGACACAAAGGCACAUCCAGAUUCGGAAUCAGAAUGAGAAUCAGAA ....((((...(((..--.....)))..............(((((((((....))))))))).......))))......(((((.............))))).... ( -23.82, z-score = -0.49, R) >droYak2.chrX 19635922 104 - 21770863 UUAUUGCCGUCAGCGA--CAACAGCUCAAUUAUUAUUGCACUUUGACCUCGGGAGGUCGGGGACACAAAGGCAUAUCCAGAUUCGGAAUCGGAAUCAGAAUCAGAA ....((((...(((..--.....)))..............(((((((((....))))))))).......))))......(((((.((.......)).))))).... ( -24.90, z-score = -0.87, R) >droEre2.scaffold_4690 17012620 98 - 18748788 UUAUUGCCAUCAGCGA--CAACAGCUCAAUUAUUAUUGCACUUUGACCUCGGGAGGUCGGGGACACAAAGGCAUGUCCAGAUACGGAAUCAGAAUCAGAA------ ....((((...(((..--.....)))..............(((((((((....))))))))).......))))..(((......))).............------ ( -22.10, z-score = -0.65, R) >droAna3.scaffold_12903 476977 77 + 802071 UUAUGGCUGUCAACGAGGCAAGAGCUCAAUUAUUAUUGCACUUUAGU--------ACCUGGGGCACAAAGG-ACACCCAGGCCGGG-------------------- ...((((((((.....))))((((..((((....))))..))))...--------..(((((.........-...)))))))))..-------------------- ( -18.20, z-score = 0.22, R) >droGri2.scaffold_15081 3654644 91 + 4274704 -UAUUUUC-UGAACUU-UAAUAAAGUCUAUUGGCACGUUUCACACACCUAAGGAUGUAAGUAGUAUUUCUGGUGAUUUCGUGUUGAAAAUAACA------------ -(((((((-...((((-.....)))).....((((((..(((((((((...)).)))...(((.....)))))))...)))))))))))))...------------ ( -16.80, z-score = -0.59, R) >consensus UUAUUGCCGUCAGCGA__CAACAGCUCAAUUAUUAUUGCACUUUGACCUCGGGAGGUCGGGGACACAAAGGCACAUCCAGAUUCGGAAUCAGAAUCAGAA______ ....((((...............((............)).(((((((((....))))))))).......))))..(((......)))................... (-13.12 = -12.63 + -0.49)
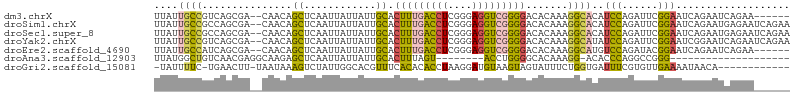
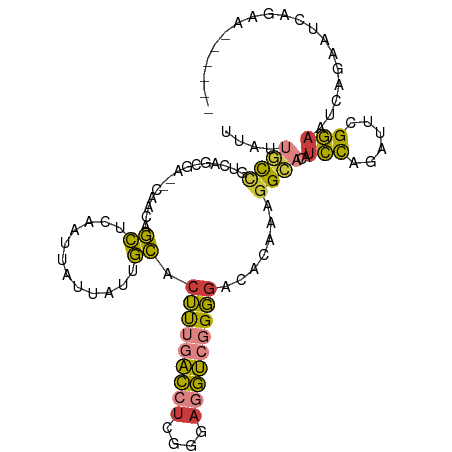
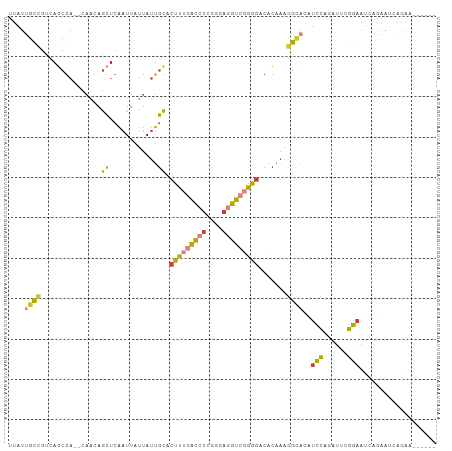
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:06 2011