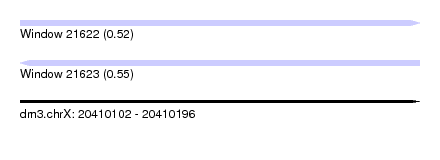
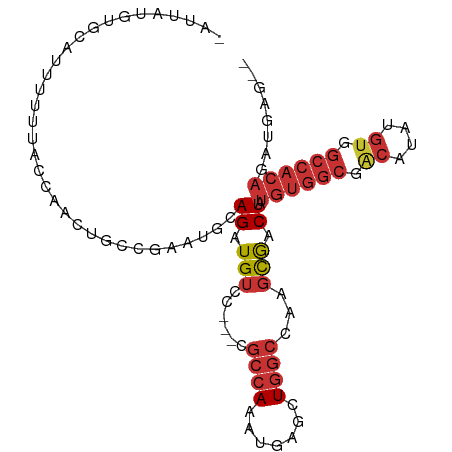
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,410,102 – 20,410,196 |
| Length | 94 |
| Max. P | 0.547228 |

| Location | 20,410,102 – 20,410,196 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 74.59 |
| Shannon entropy | 0.51637 |
| G+C content | 0.50700 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -14.33 |
| Energy contribution | -15.25 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20410102 94 + 22422827 --CUCAUCUGUGGCCACAUGGGCCGCCACAUAGUCGCUUGGCCAGCUCAUUUGGCG---GGACAUCUGCAUUCGGCAGUUGGUAAAAAAAAUGCAUAUA- --......((((((((.((((((.((((..........))))..)))))).)))).---...((.((((.....)))).))............))))..- ( -27.20, z-score = 0.10, R) >droSim1.chrX_random 5330728 95 + 5698898 --CUCAUCUGUGGCCACAUGGGCCGCCACAUAGUCGCUUGGCCAGCUCAUUUGGCG---GGACAUCUGCAUUCGGCAGUUGGUAAAAAAUGCACAUAAUC --......((((((((.((((((.((((..........))))..)))))).)))).---...((.((((.....)))).))..........))))..... ( -29.10, z-score = -0.66, R) >droSec1.super_8 2606164 95 + 3762037 --CUCAUCUGUGGCCACAUGGGCCGCCACAUAGUCGCUUGGCCAGCUCAUUUGGCG---GGACAUCUGCAUUCGGCAGUUGGUAAAAAAUGCACAUAAUC --......((((((((.((((((.((((..........))))..)))))).)))).---...((.((((.....)))).))..........))))..... ( -29.10, z-score = -0.66, R) >droYak2.chrX 19634936 83 + 21770863 --------------CUCAUUUGUGGCCACAUAGUCGCUUGGCCAGCUCAUUUGGCU---GGACAUCUGCAUUCGGCAGUUGGUAAAAAAUGCACAUAAUA --------------..(((((...((((..........((.((((((.....))))---)).)).((((.....)))).))))...)))))......... ( -21.30, z-score = -0.18, R) >droEre2.scaffold_4690 17011558 94 + 18748788 --AUCAACUGUGGCCACAUAG-CCGCCACAUAGUCGCUUGGCCAGCUCGUUUGGCG---GGACAUCUGCAUUCGGCAGUUGGUAAAAAAUGCACAUAAUA --((((((((((((((...((-(.((......)).))))))))....((....(((---(.....))))...)))))))))))................. ( -28.50, z-score = -0.72, R) >dp4.chrXL_group1e 9033479 97 + 12523060 CUCGUAUCUGUGGCCACAUAUGUCGCCACAUAGUCGCUUGGCCAGUUCAUUUGGCCAGCGGACAGCUGCAUUCAGCGGC-GCAAAAAAAUGUAUAUAA-- ........((((((.((....)).))))))..((((((.((((((.....))))))))).))).(((((.....)))))-..................-- ( -35.80, z-score = -3.08, R) >droPer1.super_22 512913 97 - 1688296 CUCGUAUCUGUGGCCACAUAUGUCGCCACAUAGUCGCUUGGCCAGUUCAUUUGGCCAGCGGACAGCUGCAUUCAGCGGC-AGAAAAAAAUGUAUAUAA-- ........((((((.((....)).))))))..((((((.((((((.....))))))))).))).(((((.....)))))-..................-- ( -35.80, z-score = -3.37, R) >droGri2.scaffold_15081 3653868 81 - 4274704 --CGUCUCUGUGGCCACAUAUGUCGCCACAUAGCUACUGGGCGAGCUCAUUUGUUG----UCCAGCUGCGCCC--UGGUUGUCCUGUAU----------- --.......(((((.((....)).)))))(((((((..(((((((((((.....))----...)))).)))))--))))))).......----------- ( -27.70, z-score = -1.44, R) >consensus __CUCAUCUGUGGCCACAUAGGCCGCCACAUAGUCGCUUGGCCAGCUCAUUUGGCG___GGACAUCUGCAUUCGGCAGUUGGUAAAAAAUGCACAUAAU_ ........((((((..........))))))..(((.....(((((.....))))).....)))..((((.....))))...................... (-14.33 = -15.25 + 0.92)

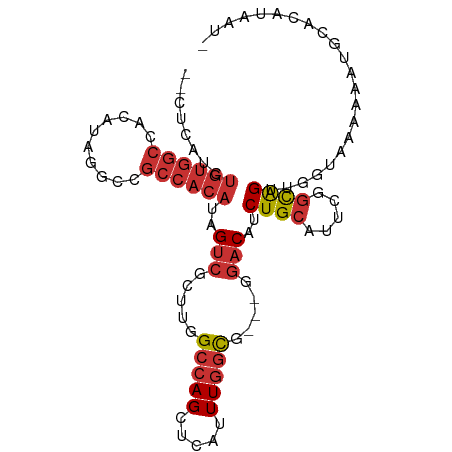
| Location | 20,410,102 – 20,410,196 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 74.59 |
| Shannon entropy | 0.51637 |
| G+C content | 0.50700 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -13.31 |
| Energy contribution | -14.75 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20410102 94 - 22422827 -UAUAUGCAUUUUUUUUACCAACUGCCGAAUGCAGAUGUCC---CGCCAAAUGAGCUGGCCAAGCGACUAUGUGGCGGCCCAUGUGGCCACAGAUGAG-- -......(((((......(((.((((.....))))(((..(---(((((.(((.(((.....)))...))).))))))..))).)))....)))))..-- ( -25.60, z-score = -0.48, R) >droSim1.chrX_random 5330728 95 - 5698898 GAUUAUGUGCAUUUUUUACCAACUGCCGAAUGCAGAUGUCC---CGCCAAAUGAGCUGGCCAAGCGACUAUGUGGCGGCCCAUGUGGCCACAGAUGAG-- .........(((((....(((.((((.....))))(((..(---(((((.(((.(((.....)))...))).))))))..))).)))....)))))..-- ( -26.80, z-score = -0.45, R) >droSec1.super_8 2606164 95 - 3762037 GAUUAUGUGCAUUUUUUACCAACUGCCGAAUGCAGAUGUCC---CGCCAAAUGAGCUGGCCAAGCGACUAUGUGGCGGCCCAUGUGGCCACAGAUGAG-- .........(((((....(((.((((.....))))(((..(---(((((.(((.(((.....)))...))).))))))..))).)))....)))))..-- ( -26.80, z-score = -0.45, R) >droYak2.chrX 19634936 83 - 21770863 UAUUAUGUGCAUUUUUUACCAACUGCCGAAUGCAGAUGUCC---AGCCAAAUGAGCUGGCCAAGCGACUAUGUGGCCACAAAUGAG-------------- .....((((.............((((.....)))).((.((---((((....).))))).)).((.((...)).))))))......-------------- ( -17.90, z-score = -0.01, R) >droEre2.scaffold_4690 17011558 94 - 18748788 UAUUAUGUGCAUUUUUUACCAACUGCCGAAUGCAGAUGUCC---CGCCAAACGAGCUGGCCAAGCGACUAUGUGGCGG-CUAUGUGGCCACAGUUGAU-- ...(((.(((((((.............))))))).)))...---.((((.......))))....(((((..(((((.(-(...)).))))))))))..-- ( -25.32, z-score = 0.11, R) >dp4.chrXL_group1e 9033479 97 - 12523060 --UUAUAUACAUUUUUUUGC-GCCGCUGAAUGCAGCUGUCCGCUGGCCAAAUGAACUGGCCAAGCGACUAUGUGGCGACAUAUGUGGCCACAGAUACGAG --..................-...((((....))))(((((((((((((.......))))).))))....((((((.((....)).)))))))))).... ( -33.50, z-score = -2.64, R) >droPer1.super_22 512913 97 + 1688296 --UUAUAUACAUUUUUUUCU-GCCGCUGAAUGCAGCUGUCCGCUGGCCAAAUGAACUGGCCAAGCGACUAUGUGGCGACAUAUGUGGCCACAGAUACGAG --..................-...((((....))))(((((((((((((.......))))).))))....((((((.((....)).)))))))))).... ( -33.50, z-score = -3.06, R) >droGri2.scaffold_15081 3653868 81 + 4274704 -----------AUACAGGACAACCA--GGGCGCAGCUGGA----CAACAAAUGAGCUCGCCCAGUAGCUAUGUGGCGACAUAUGUGGCCACAGAGACG-- -----------.(((.((....)).--(((((.((((.(.----.......).))))))))).)))(((.((((((.((....)).)))))).)).).-- ( -31.60, z-score = -3.57, R) >consensus _AUUAUGUGCAUUUUUUACCAACUGCCGAAUGCAGAUGUCC___CGCCAAAUGAGCUGGCCAAGCGACUAUGUGGCGACAUAUGUGGCCACAGAUGAG__ .....................................(((.....((((.......)))).....)))..((((((.((....)).))))))........ (-13.31 = -14.75 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:04:04 2011