| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,337,033 – 20,337,127 |
| Length | 94 |
| Max. P | 0.816876 |

| Location | 20,337,033 – 20,337,127 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 56.29 |
| Shannon entropy | 0.74799 |
| G+C content | 0.33450 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -5.80 |
| Energy contribution | -5.44 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
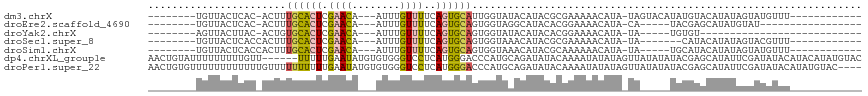
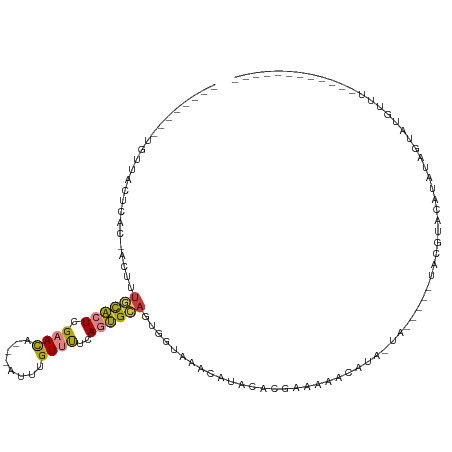
>dm3.chrX 20337033 94 + 22422827 --------UGUUACUCAC-ACUUUGCACUCGAACA---AUUUGUUUUCAGUGCAUUGGUAUACAUACGCGAAAAACAUA-UAGUACAUAUGUACAUAUAGUAUGUUU------------ --------..........-(((.((((((.((((.---....))))..))))))..)))..((((((.......(((((-(.....)))))).......))))))..------------ ( -19.94, z-score = -1.21, R) >droEre2.scaffold_4690 16941672 84 + 18748788 --------UGUUACUCAC-ACUUUGCACUCGAACA---AUUUGUUUUCAGUGCAGUGGUAGGCAUACACGGAAAACAUA-CA-----UACGAGCAUAUGUAU----------------- --------((((((.(((-....((((((.((((.---....))))..)))))))))))).)))............(((-((-----((......)))))))----------------- ( -17.40, z-score = -0.73, R) >droYak2.chrX 19562492 74 + 21770863 --------AGUUACUUAC-ACUGUGCACUCGAACA---AUUUGUUUUCAGUGCAGUGGUAUACAUACACGGAAAACAUA-UA-----UGUGU--------------------------- --------.........(-((((..(....((((.---....))))...)..)))))..(((((((...(.....)...-))-----)))))--------------------------- ( -15.30, z-score = -0.08, R) >droSec1.super_8 2531715 88 + 3762037 --------UGUUACUCACCACUUUGCACUCGAACA---AUUUGUUUUCAGUGCAGUGGUAAACAUACGCGAAAAACAUA-UA-------CAUACAUAUAGUACGUUU------------ --------........((((((..(((((.((((.---....))))..)))))))))))((((.(((............-..-------..........))).))))------------ ( -16.96, z-score = -1.19, R) >droSim1.chrX 15594523 90 + 17042790 --------UGUUACUCACCACUUUGCACUCGAACA---AUUUGUUUUCAGUGCAGUGGUAAACAUACGCAAAAAACAUA-UA-----UGCAUACAUAUAGUAUGUUU------------ --------........((((((..(((((.((((.---....))))..)))))))))))(((((((((.......).((-((-----((....))))))))))))))------------ ( -24.00, z-score = -3.27, R) >dp4.chrXL_group1e 8946328 113 + 12523060 AACUGUAUUUUUUUUUGUU------UUUUUGAAUAUGUGUGGGUCCUCAUGGGACCCAUGCAGAUAUACAAAAUAUAUAGUUAUAUAUACGAGCAUAUUCGAUAUACAUACAUAUGUAC ((((((((....((((((.------..........(((((((((((.....))))))))))).....)))))).)))))))).(((((.((((....)))))))))............. ( -30.39, z-score = -3.36, R) >droPer1.super_22 424333 115 - 1688296 AACUGUGUUUUUUUUUUUUGUUUUUUUUUUGAAUAUGUGUGGGUCCUCAUGGGACCCAUGCAGAUAUACAAAAUAUAUAGUUAUAUAUACGAGCAUAUUCGAUAUACAUAUGUAC---- .(((((((..........(((((...(((((.((((((((((((((.....)))))))))))..))).)))))((((((....)))))).)))))..........))))).))..---- ( -29.25, z-score = -3.26, R) >consensus ________UGUUACUCAC_ACUUUGCACUCGAACA___AUUUGUUUUCAGUGCAGUGGUAAACAUACACGAAAAACAUA_UA_____UACGUACAUAUAGUAUGUUU____________ .......................((((((.((((........))))..))))))................................................................. ( -5.80 = -5.44 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:53 2011