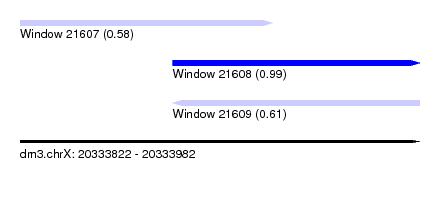
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,333,822 – 20,333,982 |
| Length | 160 |
| Max. P | 0.994303 |

| Location | 20,333,822 – 20,333,923 |
|---|---|
| Length | 101 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.99 |
| Shannon entropy | 0.40776 |
| G+C content | 0.63389 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -21.18 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
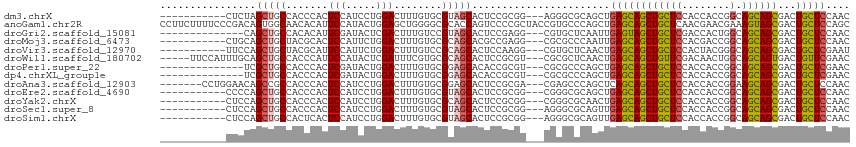
>dm3.chrX 20333822 101 + 22422827 -----------CUCUAGCUGCCACCCACUCCAUCCUGGACUUUGUGCGUAGCACUCCGCGG---AGGGCGCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAAC -----------....((((((...((.((((....((((....((((...)))))))).))---)))).))))))(((((((((((((......)).))))))...))))).... ( -40.60, z-score = -1.57, R) >anoGam1.chr2R 28959143 115 - 62725911 CCUUCUUUUCCCGACAGUGGCAACACAUUCCAUACUGGAGCUGGGGCGCACCAGUCCCCGCUACCGUGCCCAGCUGAGCAGCUGCUCAACGAACGAAAGUAGCGACUGCUCCAGC ................(((....)))........(((((((.(((((......)))))(((((((((.....(.(((((....))))).)..)))...))))))...))))))). ( -43.70, z-score = -2.81, R) >droGri2.scaffold_15081 3581417 98 - 4274704 --------------CAGCUGCCACACAUUCGAUACUCGACUUUGUCCGUAGCACUCCGAGG---CGUGCUCAAUUGAGUAGCUGCUCGACCACUGGCAGCAGCGACUGCUCCAAC --------------..(((((((.....(((.....)))....(((.(((((((((.(((.---....)))....)))).)))))..)))...)))))))(((....)))..... ( -30.70, z-score = -0.96, R) >droMoj3.scaffold_6473 4030442 101 + 16943266 -----------CUGCAGCUGCUACGCACUCCAUUCUGGACUUUGUGCGCAGCACGCCGAGG---CGCGCCCAAUUGAGCAGCUGCUCCACGACCGGCAGCAGCGACUGCUCCAAC -----------..(((((((((.((((((((.....)))....)))))..((.(((....)---)).)).......))))))))).........((.(((((...)))))))... ( -42.50, z-score = -2.08, R) >droVir3.scaffold_12970 3173350 101 + 11907090 -----------UUCCAGCUGCUACGCAUUCCAUUCUGGACUUUGUCCGCAGCACUCCAAGG---CGUGCUCAACUGAGCAGCUGCUCCACUACGGGCAGCAGCGACUGCUCGAAU -----------...((((((((..............((((...))))(.(((((.(....)---.)))))).....))))))))........(((((((......)))))))... ( -36.50, z-score = -1.76, R) >droWil1.scaffold_180702 1553609 107 - 4511350 -----UUCCAUUUGCAGCUGCCACCCAUUCCAUACUCGAUUUCGUGCGCAGCACUCCGCGU---CGCGCUCAACUGAGCAGCUGUUCGACAACUGGCAGCAGUGACUGUUCGAAC -----........(((((((((((....((.......))....))).))))).......((---(((((((....)))).(((((..(....)..))))).))))))))...... ( -32.60, z-score = -1.03, R) >droPer1.super_22 421552 98 - 1688296 --------------UCGCUGCCACCCACUCGAUACUGGACUUUGUGCGGAGCACACCGCGU---CGCGCCCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCGAAC --------------..((((....(((........))).....(((((..((.....))..---))))).))))((((((((((((((......)).))))))...))))))... ( -36.90, z-score = -1.36, R) >dp4.chrXL_group1e 8943559 98 + 12523060 --------------UCGCUGCCACCCACUCGAUACUGGACUUUGUGCGGAGCACACCGCGU---CGCGCCCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCGAAC --------------..((((....(((........))).....(((((..((.....))..---))))).))))((((((((((((((......)).))))))...))))))... ( -36.90, z-score = -1.36, R) >droAna3.scaffold_12903 384918 105 - 802071 -------CCUGGAACAGCCGCCACCCACUCCAUCCUGGACUUUGUGCGGAGCACUCCGCGA---CGAGCCCAGCUCAGCAGCUGCUCCACCACCGGAAGCAGCGACUGCUCCAAC -------..((((.(((.(...............((((.(((..((((((....)))))).---.))).)))).......((((((((......)).))))))).))).)))).. ( -38.50, z-score = -2.42, R) >droEre2.scaffold_4690 16938933 101 + 18748788 -----------CCCCAGCUGCCACCCACUCCAUCCUGGACUUUGUGCGUAGCACUCCGCGG---CGGGCGCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAAC -----------.....((((((((....(((.....)))....))).))))).......((---.(((.((((((....))))))))).))...((.(((((...)))))))... ( -39.00, z-score = -1.01, R) >droYak2.chrX 19559631 101 + 21770863 -----------CUCCAGCUGCCACCCACUCCAUCCUGGACUUUGUGCGCAGCACUCCGCGG---CGGGCGCAACUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAAC -----------.....((((((((....(((.....)))....))).))))).....(((.---....)))....(((((((((((((......)).))))))...))))).... ( -37.40, z-score = -1.07, R) >droSec1.super_8 2528248 101 + 3762037 -----------CUCCAGCUGCCACCCACUCCAUCCUGGACUUUGUGCGUAGCACUCCGCGG---AGGGCGCAGUUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAAC -----------.....((((((((....(((.....)))....))).))))).(((....)---))((.(((((((....((((((........))))))..))))))).))... ( -40.40, z-score = -1.69, R) >droSim1.chrX 15588950 101 + 17042790 -----------CUCCAGCUGCCACUCACUCCAUCCUGGACUUUGUGCGUAGCACUCCGCGG---AGGGCGCAGUUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAAC -----------.....((((((((....(((.....)))....))).))))).(((....)---))((.(((((((....((((((........))))))..))))))).))... ( -41.10, z-score = -1.82, R) >consensus ___________CUCCAGCUGCCACCCACUCCAUCCUGGACUUUGUGCGCAGCACUCCGCGG___CGGGCCCAGCUGAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAAC ................(((((.......(((.....)))........))))).......................((((((((((((........).))))))...))))).... (-21.18 = -21.42 + 0.24)
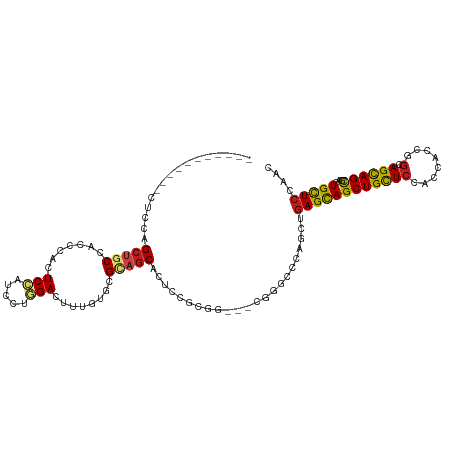
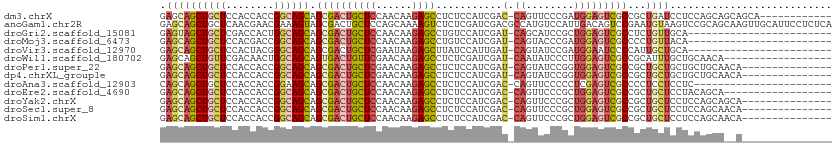
| Location | 20,333,883 – 20,333,982 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.96 |
| Shannon entropy | 0.45137 |
| G+C content | 0.60827 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -19.25 |
| Energy contribution | -19.51 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
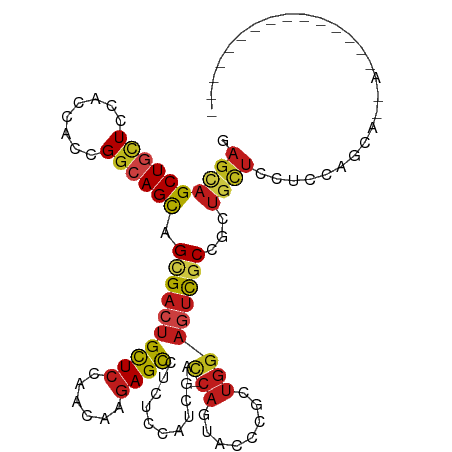
>dm3.chrX 20333883 99 + 22422827 GAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGAC-CAGUUCCCGAUGGAGUCGCCGCUGAUCCUCCAGCAGCAGCA------------ ..((.((((((((......))((((.(((((.((((......))))...(((((((..-.......)))))))))))).)))).......)))))).)).------------ ( -41.70, z-score = -4.01, R) >anoGam1.chr2R 28959218 112 - 62725911 GAGCAGCUGCUCAACGAACGAAAGUAGCGACUGCUCCAGCAAAAGUCUCUCGAUCGACGCCAUGUCCAUUGACAGUCCGAAUGUAAGUCCGCAGCAAGUUGCAUUCCUCUCA ((((((.((((......((....)))))).))))))........(.((.(((((.(((.....))).))))).)).).((((((((.(........).))))))))...... ( -29.40, z-score = -0.72, R) >droGri2.scaffold_15081 3581475 87 - 4274704 GAGUAGCUGCUCGACCACUGGCAGCAGCGACUGCUCCAACAAGAGCCUGUCCAUCGAU-CAGCAUCCGCUGGAGUCGCCUCUGUUGCA------------------------ (((..((((((........)))))).((((((((((......))))...........(-((((....)))))))))))))).......------------------------ ( -30.80, z-score = -0.74, R) >droMoj3.scaffold_6473 4030503 87 + 16943266 GAGCAGCUGCUCCACGACCGGCAGCAGCGACUGCUCCAACAAGAGCCUGUCCAUCGAU-CAGUACCCGAUGGAGUCGCCCCUGUUACA------------------------ .((((((((((........)))))).(((((.((((......))))...(((((((..-.......))))))))))))...))))...------------------------ ( -32.90, z-score = -3.09, R) >droVir3.scaffold_12970 3173411 87 + 11907090 GAGCAGCUGCUCCACUACGGGCAGCAGCGACUGCUCGAAUAAGAGCUUAUCCAUUGAU-CAGUAUCCGAUGGAAUCCCCAUUGCUGCA------------------------ ..(((((.((((.....(((((((......))))))).....))))...(((((((..-.......))))))).........))))).------------------------ ( -32.70, z-score = -2.96, R) >droWil1.scaffold_180702 1553676 93 - 4511350 GAGCAGCUGUUCGACAACUGGCAGCAGUGACUGUUCGAACAAGAGCCUCUCGAUCGAU-CAAUAUCCCUUGGAGUCGCCGCAUUUGCUGCAACA------------------ ..((((((((((((((((((....))))...)).))))))).(.((..(((.(..(((-....)))...).)))..)))......)))))....------------------ ( -26.80, z-score = -0.28, R) >droPer1.super_22 421610 96 - 1688296 GAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCGAACAAGAGCCUCUCCAUCGAU-CAGUAUCCGGUGGAGUCGCCGCUGCUGCUGCUGCAACA--------------- ..(((((.((.........((((((.(((((.((((......))))...(((((((..-.......)))))))))))).)))))))).)))))....--------------- ( -44.60, z-score = -3.37, R) >dp4.chrXL_group1e 8943617 96 + 12523060 GAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCGAACAAGAGCCUCUCCAUCGAU-CAGUAUCCGGUGGAGUCGCCGCUGCUGCUGCUGCAACA--------------- ..(((((.((.........((((((.(((((.((((......))))...(((((((..-.......)))))))))))).)))))))).)))))....--------------- ( -44.60, z-score = -3.37, R) >droAna3.scaffold_12903 384983 88 - 802071 CAGCAGCUGCUCCACCACCGGAAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGAC-CAGUUCCCCCUCGAGUCGCCCCUCCUCCUC----------------------- ..((.((((((((......)).))))))(((.((((......)))).......((((.-..........)))))))))...........----------------------- ( -23.90, z-score = -2.42, R) >droEre2.scaffold_4690 16938994 93 + 18748788 GAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGAC-CAGUUCCCGCUGGAGUCGCCGCUGCUCCUACAGCA------------------ (((((((((((((......)).))))((((((((((......))))...........(-((((....))))))))))).)))))))........------------------ ( -39.10, z-score = -3.84, R) >droYak2.chrX 19559692 96 + 21770863 GAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGAC-CAGUUCCCGCUGGAGUCGCCGCUGCUCCUCCAGCAGCA--------------- .....((((((........((((((.((((((((((......))))...........(-((((....))))))))))).)))))).....)))))).--------------- ( -40.92, z-score = -3.72, R) >droSec1.super_8 2528309 96 + 3762037 GAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGAC-CAGUUCCCGCUGGAGUCGCCGCUGCUCCUCCAGCAACA--------------- (((((((((((((......)).))))((((((((((......))))...........(-((((....))))))))))).)))))))...........--------------- ( -39.10, z-score = -3.85, R) >droSim1.chrX 15589011 96 + 17042790 GAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGAC-CAGUUCCCGCUGGAGUCGCCGCUGCUCCUCCAGCAACA--------------- (((((((((((((......)).))))((((((((((......))))...........(-((((....))))))))))).)))))))...........--------------- ( -39.10, z-score = -3.85, R) >consensus GAGCAGCUGCUCCACCACCGGCAGCAGCGACUGCUCCAACAAGAGCCUCUCCAUCGAC_CAGUACCCGCUGGAGUCGCCGCUGCUCCUCCAGCA__A_______________ .((((((((((........)))))).((((((((((......))))..................((....))))))))...))))........................... (-19.25 = -19.51 + 0.26)
| Location | 20,333,883 – 20,333,982 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Shannon entropy | 0.45137 |
| G+C content | 0.60827 |
| Mean single sequence MFE | -42.24 |
| Consensus MFE | -17.54 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
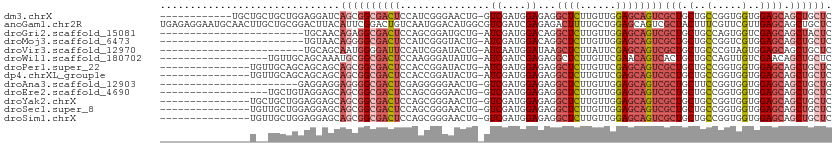
>dm3.chrX 20333883 99 - 22422827 ------------UGCUGCUGCUGGAGGAUCAGCGGCGACUCCAUCGGGAACUG-GUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUC ------------.((((((((((..((..((((((((((((((((((......-.))))))))...(((((....))))).))))))))))))..))))..))))))..... ( -50.20, z-score = -2.82, R) >anoGam1.chr2R 28959218 112 + 62725911 UGAGAGGAAUGCAACUUGCUGCGGACUUACAUUCGGACUGUCAAUGGACAUGGCGUCGAUCGAGAGACUUUUGCUGGAGCAGUCGCUACUUUCGUUCGUUGAGCAGCUGCUC ((.(((...((((......))))..))).))..(((.((((((((((((.(((((.(..((....))....(((....)))).))))).....))))))))).))))))... ( -30.70, z-score = 1.18, R) >droGri2.scaffold_15081 3581475 87 + 4274704 ------------------------UGCAACAGAGGCGACUCCAGCGGAUGCUG-AUCGAUGGACAGGCUCUUGUUGGAGCAGUCGCUGCUGCCAGUGGUCGAGCAGCUACUC ------------------------.(((.(((.(((..(((((((((..(((.-.((....))..)))..)))))))))..))).))).))).((((((......)))))). ( -34.80, z-score = -0.78, R) >droMoj3.scaffold_6473 4030503 87 - 16943266 ------------------------UGUAACAGGGGCGACUCCAUCGGGUACUG-AUCGAUGGACAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUCGUGGAGCAGCUGCUC ------------------------..((((((((((...((((((((......-.))))))))...))))))))))(((((...((((((.(((....)))))))))))))) ( -41.30, z-score = -2.93, R) >droVir3.scaffold_12970 3173411 87 - 11907090 ------------------------UGCAGCAAUGGGGAUUCCAUCGGAUACUG-AUCAAUGGAUAAGCUCUUAUUCGAGCAGUCGCUGCUGCCCGUAGUGGAGCAGCUGCUC ------------------------.(((((.........(((((..(((....-))).)))))...(((((....((.(((((....))))).))....))))).))))).. ( -31.70, z-score = -1.41, R) >droWil1.scaffold_180702 1553676 93 + 4511350 ------------------UGUUGCAGCAAAUGCGGCGACUCCAAGGGAUAUUG-AUCGAUCGAGAGGCUCUUGUUCGAACAGUCACUGCUGCCAGUUGUCGAACAGCUGCUC ------------------....(((((.....((((((((...((((..((((-.((((.((((.....)))).)))).))))..)).))...))))))))....))))).. ( -31.50, z-score = -0.99, R) >droPer1.super_22 421610 96 + 1688296 ---------------UGUUGCAGCAGCAGCAGCGGCGACUCCACCGGAUACUG-AUCGAUGGAGAGGCUCUUGUUCGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUC ---------------....(((((.(((((((((((..(((((.(((......-.))).)))))..((((......)))).)))))))))))..((......)).))))).. ( -46.20, z-score = -1.97, R) >dp4.chrXL_group1e 8943617 96 - 12523060 ---------------UGUUGCAGCAGCAGCAGCGGCGACUCCACCGGAUACUG-AUCGAUGGAGAGGCUCUUGUUCGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUC ---------------....(((((.(((((((((((..(((((.(((......-.))).)))))..((((......)))).)))))))))))..((......)).))))).. ( -46.20, z-score = -1.97, R) >droAna3.scaffold_12903 384983 88 + 802071 -----------------------GAGGAGGAGGGGCGACUCGAGGGGGAACUG-GUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUUCCGGUGGUGGAGCAGCUGCUG -----------------------.....((((.((((((((((.((....)).-.)))).......(((((....))))).)))))).))))(((..((......))..))) ( -33.30, z-score = -0.62, R) >droEre2.scaffold_4690 16938994 93 - 18748788 ------------------UGCUGUAGGAGCAGCGGCGACUCCAGCGGGAACUG-GUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUC ------------------.((..(.(((((((((((..(((((((((((.((.-.((....))..)).)))))))))))..))))))))).)).)..)).((((....)))) ( -48.60, z-score = -3.09, R) >droYak2.chrX 19559692 96 - 21770863 ---------------UGCUGCUGGAGGAGCAGCGGCGACUCCAGCGGGAACUG-GUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUC ---------------.((..((((...(((((((((..(((((((((((.((.-.((....))..)).)))))))))))..))))))))).))))..)).((((....)))) ( -53.60, z-score = -3.81, R) >droSec1.super_8 2528309 96 - 3762037 ---------------UGUUGCUGGAGGAGCAGCGGCGACUCCAGCGGGAACUG-GUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUC ---------------..(..((((...(((((((((..(((((((((((.((.-.((....))..)).)))))))))))..))))))))).))))..)..((((....)))) ( -50.50, z-score = -3.34, R) >droSim1.chrX 15589011 96 - 17042790 ---------------UGUUGCUGGAGGAGCAGCGGCGACUCCAGCGGGAACUG-GUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUC ---------------..(..((((...(((((((((..(((((((((((.((.-.((....))..)).)))))))))))..))))))))).))))..)..((((....)))) ( -50.50, z-score = -3.34, R) >consensus _______________U__UGCUGGAGGAGCAGCGGCGACUCCAGCGGGAACUG_GUCGAUGGAGAGGCUCUUGUUGGAGCAGUCGCUGCUGCCGGUGGUGGAGCAGCUGCUC ..............................((((((((((...............((....))...((((......))))))))(((.(..(.....)..)))).)))))). (-17.54 = -18.40 + 0.86)
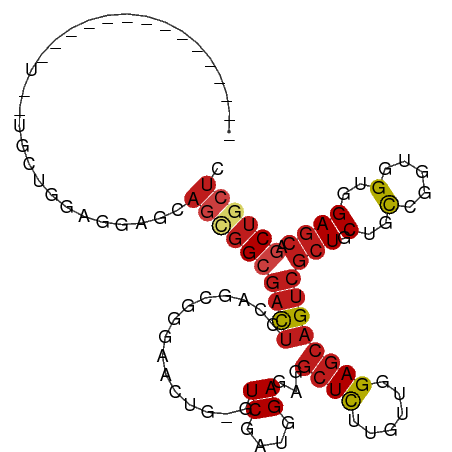
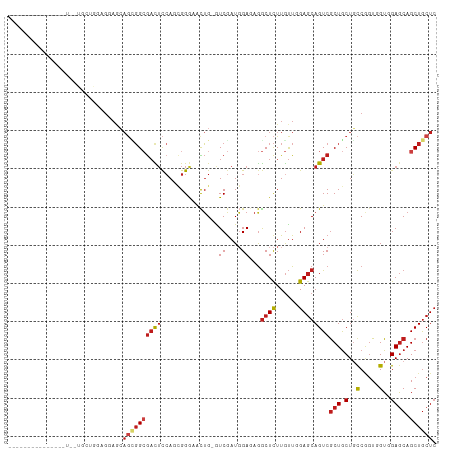
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:52 2011