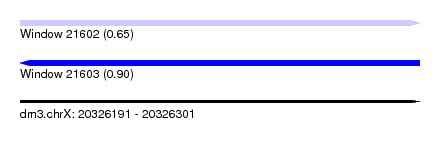
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,326,191 – 20,326,301 |
| Length | 110 |
| Max. P | 0.904945 |

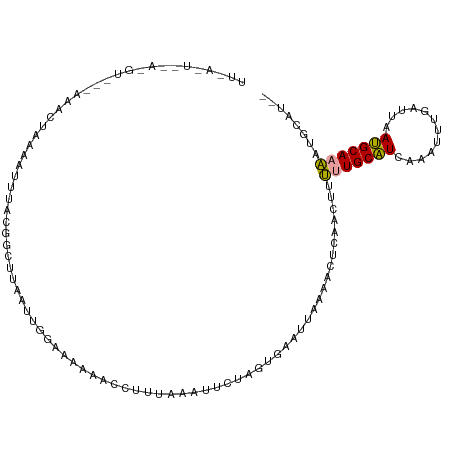
| Location | 20,326,191 – 20,326,301 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 47.31 |
| Shannon entropy | 0.75487 |
| G+C content | 0.29170 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -4.42 |
| Energy contribution | -3.87 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20326191 110 + 22422827 UUUACUUUAGGUCGCAAUCUAAACUUUCUGAUUCAAUUUGCAAAAACUUUUCAAUUCUAGUGAAUUGAAAAUCAACUUUUUGCAUUCAAUUUGAUUAAUGCAAAAUGCAU-- .....(((((((....))))))).....(((((.((((((((((((.((((((((((....)))))))))).....))))))))...)))).)))))((((.....))))-- ( -22.20, z-score = -2.58, R) >droYak2.chrX 19552568 110 + 21770863 UUAAUUAAAAGUUUGAAGUUGAAACUUUCCGCUUAAAAGGGAAAAACGUUUAAUUUCUAGAGAAUUAUAACUCAACUUUUUGCAUCAAAUUCUUUUAAUGCAAAGUGCAU-- .......((((((.(((((((((..(((((.((....)))))))....)))))))))..(((........)))))))))((((((.(((....))).)))))).......-- ( -20.90, z-score = -1.61, R) >droGri2.scaffold_14830 1681272 92 + 6267026 -----------------AGUAGAUUUUACGGCGUAAUUGAAAUAAAUCUCAUACCCGUAAUGCUUUUAAGC---AAUUGAUGCUUCAGACAUGAAAAAGGCAACCGAAACAA -----------------........((((((.(((..(((........))))))))))))(((((((..((---(.....)))((((....))))))))))).......... ( -18.40, z-score = -1.69, R) >consensus UU_A_U__A_GU___AAACUAAAAUUUACGGCUUAAUUGGAAAAAACCUUUAAAUUCUAGUGAAUUAAAACUCAACUUUUUGCAUCAAAUUUGAUUAAUGCAAAAUGCAU__ ..............................................................................(((((((............)))))))........ ( -4.42 = -3.87 + -0.55)

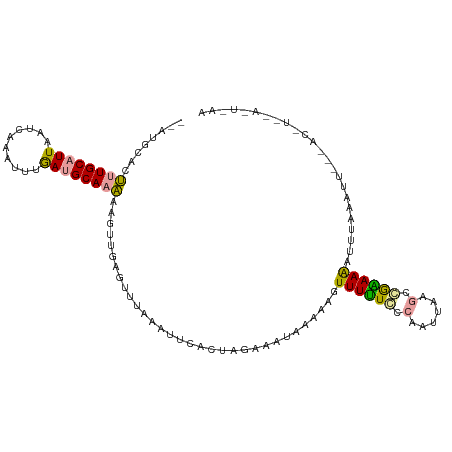
| Location | 20,326,191 – 20,326,301 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 47.31 |
| Shannon entropy | 0.75487 |
| G+C content | 0.29170 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -5.94 |
| Energy contribution | -5.40 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20326191 110 - 22422827 --AUGCAUUUUGCAUUAAUCAAAUUGAAUGCAAAAAGUUGAUUUUCAAUUCACUAGAAUUGAAAAGUUUUUGCAAAUUGAAUCAGAAAGUUUAGAUUGCGACCUAAAGUAAA --.(((.(((((((((..........)))))))))......((((((((((....))))))))))....((((((.((((((......)))))).))))))......))).. ( -24.60, z-score = -2.03, R) >droYak2.chrX 19552568 110 - 21770863 --AUGCACUUUGCAUUAAAAGAAUUUGAUGCAAAAAGUUGAGUUAUAAUUCUCUAGAAAUUAAACGUUUUUCCCUUUUAAGCGGAAAGUUUCAACUUCAAACUUUUAAUUAA --......(((((((((((....)))))))))))((((((((((.((((((....).))))))))..((((((((....)).))))))..)))))))............... ( -21.80, z-score = -2.08, R) >droGri2.scaffold_14830 1681272 92 - 6267026 UUGUUUCGGUUGCCUUUUUCAUGUCUGAAGCAUCAAUU---GCUUAAAAGCAUUACGGGUAUGAGAUUUAUUUCAAUUACGCCGUAAAAUCUACU----------------- .((((((((.((.......))...)))))))).....(---(((....))))((((((((((((((....)))))..))).))))))........----------------- ( -20.60, z-score = -2.00, R) >consensus __AUGCACUUUGCAUUAAUCAAAUUUGAUGCAAAAAGUUGAGUUUAAAUUCACUAGAAAUAAAAAGUUUUUCCCAAUUAAGCCGAAAAAUUUAAAUU___AC_U__A_U_AA ........((((((((..........))))))))................................((((((.(......).))))))........................ ( -5.94 = -5.40 + -0.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:47 2011