
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,312,159 – 20,312,253 |
| Length | 94 |
| Max. P | 0.936147 |

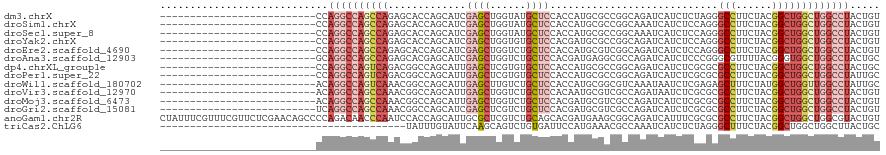
| Location | 20,312,159 – 20,312,253 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.91 |
| Shannon entropy | 0.40841 |
| G+C content | 0.61102 |
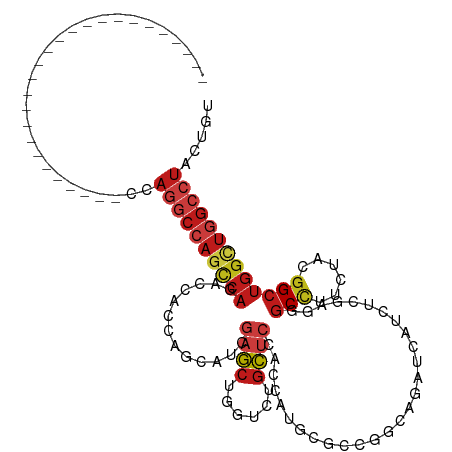
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -23.34 |
| Energy contribution | -24.39 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20312159 94 + 22422827 --------------------------CCAGGCCAGCCAGAGCACCAGCAUCGAGCUGGUAUGCUCCACCAUGCGCCGGCAGAUCAUCUCUAGGGCCUUCUACGGCUGGCUGGCCUACUGU --------------------------..((((((((((((((((((((.....))))))..))))........((((..(((....((....))...))).))))))))))))))..... ( -45.20, z-score = -2.93, R) >droSim1.chrX 15576657 94 + 17042790 --------------------------CCAGGCCAGCCAGAGCACCAGCAUCGAGCUGGUAUGCUCCACCAUGCGCCGGCAAAUCAUCUCCAGGGCCUUCUACGGCUGGCUGGCCUACUGU --------------------------..((((((((((((((((((((.....))))))..))))........((((........((....))........))))))))))))))..... ( -42.69, z-score = -2.65, R) >droSec1.super_8 2506250 94 + 3762037 --------------------------CCAGGCCAGCCAGAGCACCAGCAUCGAGCUGGUAUGCUCCACCAUGCGCCGGCAAAUCAUCUCCAGGGCCUUCUACGGCUGGCUGGCCUACUGU --------------------------..((((((((((((((((((((.....))))))..))))........((((........((....))........))))))))))))))..... ( -42.69, z-score = -2.65, R) >droYak2.chrX 19539425 94 + 21770863 --------------------------CCAGGCCAGCCAGAGCACCAGCAUCGAGCUGGUGUGCUCCACGAUGCGCCGGCAGAUCAUCUCCAGGGCCUUCUACGGCUGGCUGGCCUACUGU --------------------------..((((((((((..((((((((.....))))))))((........))((((..(((....((....))...))).))))))))))))))..... ( -45.80, z-score = -2.47, R) >droEre2.scaffold_4690 16919835 94 + 18748788 --------------------------CCAGGCCAGCCAGAGCACCAGCAUCGAGCUGGUCUGCUCCACCAUGCGUCGGCAGAUCAUCUCCAGGGCCUUCUACGGCUGGCUGGCCUACUGU --------------------------..(((((((((.((((....)).))(((.((((((((.(.((.....)).))))))))).)))...((((......)))))))))))))..... ( -42.90, z-score = -2.30, R) >droAna3.scaffold_12903 364987 94 - 802071 --------------------------GCAGGCCAGCCAGAGCACGAGCAUCGAGCUGGUCUGCUCCACGAUGAGGCGCCAGAUCAUCUCCCGGGCGUUUUACGGGUGGCUGGCCUACUGC --------------------------..((((((((((((((((.(((.....))).))..))))..((.(((((((((.((.....))...)))))))))))..))))))))))..... ( -44.80, z-score = -2.31, R) >dp4.chrXL_group1e 8925113 94 + 12523060 --------------------------CCAGGCCAGUCAGACGGCCAGCAUUGAGCUCGUGUGCUCCACCAUGCGCCGGCAGAUCAUCUCGCGCGCCUUCUACGGCUGGCUGGCCUACUGC --------------------------..((((((((((..((((..((((.((((......))))....))))))))((((.....)).))..(((......)))))))))))))..... ( -39.70, z-score = -1.20, R) >droPer1.super_22 403039 94 - 1688296 --------------------------CCAGGCCAGUCAGACGGCCAGCAUUGAGCUCGUGUGCUCCACCAUGCGCCGGCAGAUCAUCUCGCGCGCCUUCUACGGCUGGCUGGCCUAUUGC --------------------------..((((((((((..((((..((((.((((......))))....))))))))((((.....)).))..(((......)))))))))))))..... ( -39.70, z-score = -1.32, R) >droWil1.scaffold_180702 1537867 94 - 4511350 --------------------------ACAGGCCAGUCAAACGGCCAGCAUUGAGCUUGUCUGCUCCACCAUGCGGCGUCAAAUAAUCUCGAGAGCUUUCUAUGGCUGGUUGGCCUAUUGC --------------------------..((((((..(.....(((.((((.((((......))))....)))))))................((((......)))))..))))))..... ( -31.60, z-score = -1.15, R) >droVir3.scaffold_12970 3147992 94 + 11907090 --------------------------ACAGGCCAGCCAAACGGCCAGCAUUGAGCUGGUCUGCUCCACAAUGCGUCGCCAGAUAAUCUCGCGCGCCUUCUACGGCUGGCUGGCCUACUGU --------------------------..((((((((((...(((((((.....))))))).(((.......(((.(((.((.....)).)))))).......)))))))))))))..... ( -45.54, z-score = -4.23, R) >droMoj3.scaffold_6473 4001648 94 + 16943266 --------------------------ACAGGCCAGCCAAACGGCCAGCAUUGAGCUGGUCUGCUCCACGAUGCGUCGCCAGAUCAUCUCGCGCGCCUUCUACGGCUGGCUGGCCUACUGU --------------------------..((((((((((...(((((((.....))))))).(((....((.(((.(((.((.....)).))))))..))...)))))))))))))..... ( -46.00, z-score = -3.86, R) >droGri2.scaffold_15081 3565247 94 - 4274704 --------------------------UCAGGCCAGCCAAACGGCCAGCAUCGAGCUCGUCUGCUCCACGAUGCGUCGCCAGAUCAUCUCGCGCGCCUUCUACGGCUGGCUGGCCUACUGU --------------------------..((((((((((...(((..(((((((((......))))...)))))...)))..............(((......)))))))))))))..... ( -43.40, z-score = -3.58, R) >anoGam1.chr2R 28959350 120 - 62725911 CUAUUUCGUUUCGUUCUCGAACAGCCCCAGACAACCCAAUCCACCAGCAUUGCGCUCGUCUGCAGCACGAUGAAGCGGCAGAUCAUUUCGCGCGCCUUCUACGGCUGGCUGGCGUACUGU ......(((((((....))))(((((.........................((((..((((((.((........)).))))))......))))(((......))).))))))))...... ( -32.10, z-score = 0.59, R) >triCas2.ChLG6 8072506 79 + 13544221 -----------------------------------------UAUUUGUAUUCAAGCAGUCUGUGAUUCCAUGAAACGCCAAAUCAUCUCUAGGGCUUUCUACGGCUGGCUGGCUUACUGC -----------------------------------------.............(((((..((........((((.(((.............)))))))..(((....)))))..))))) ( -14.72, z-score = 0.87, R) >consensus __________________________CCAGGCCAGCCAGACCACCAGCAUCGAGCUGGUCUGCUCCACCAUGCGCCGGCAGAUCAUCUCGAGGGCCUUCUACGGCUGGCUGGCCUACUGU ............................((((((((((.............((((......))))............................(((......)))))))))))))..... (-23.34 = -24.39 + 1.05)
| Location | 20,312,159 – 20,312,253 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Shannon entropy | 0.40841 |
| G+C content | 0.61102 |
| Mean single sequence MFE | -42.42 |
| Consensus MFE | -20.46 |
| Energy contribution | -21.28 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20312159 94 - 22422827 ACAGUAGGCCAGCCAGCCGUAGAAGGCCCUAGAGAUGAUCUGCCGGCGCAUGGUGGAGCAUACCAGCUCGAUGCUGGUGCUCUGGCUGGCCUGG-------------------------- ....((((((((((.(((((.(..(((...(((.....))))))..)..)))))(((((..((((((.....))))))))))))))))))))).-------------------------- ( -49.40, z-score = -3.28, R) >droSim1.chrX 15576657 94 - 17042790 ACAGUAGGCCAGCCAGCCGUAGAAGGCCCUGGAGAUGAUUUGCCGGCGCAUGGUGGAGCAUACCAGCUCGAUGCUGGUGCUCUGGCUGGCCUGG-------------------------- ....((((((((((.(((((.....((.((((..(....)..)))).)))))))(((((..((((((.....))))))))))))))))))))).-------------------------- ( -48.30, z-score = -2.98, R) >droSec1.super_8 2506250 94 - 3762037 ACAGUAGGCCAGCCAGCCGUAGAAGGCCCUGGAGAUGAUUUGCCGGCGCAUGGUGGAGCAUACCAGCUCGAUGCUGGUGCUCUGGCUGGCCUGG-------------------------- ....((((((((((.(((((.....((.((((..(....)..)))).)))))))(((((..((((((.....))))))))))))))))))))).-------------------------- ( -48.30, z-score = -2.98, R) >droYak2.chrX 19539425 94 - 21770863 ACAGUAGGCCAGCCAGCCGUAGAAGGCCCUGGAGAUGAUCUGCCGGCGCAUCGUGGAGCACACCAGCUCGAUGCUGGUGCUCUGGCUGGCCUGG-------------------------- ....(((((((((((((((..(...((.((((((.....)).)))).)).)..)))....(((((((.....)))))))..)))))))))))).-------------------------- ( -47.80, z-score = -2.46, R) >droEre2.scaffold_4690 16919835 94 - 18748788 ACAGUAGGCCAGCCAGCCGUAGAAGGCCCUGGAGAUGAUCUGCCGACGCAUGGUGGAGCAGACCAGCUCGAUGCUGGUGCUCUGGCUGGCCUGG-------------------------- ....((((((((((.(((((.(..(((...(((.....))))))..)..)))))(((((..((((((.....))))))))))))))))))))).-------------------------- ( -49.10, z-score = -3.35, R) >droAna3.scaffold_12903 364987 94 + 802071 GCAGUAGGCCAGCCACCCGUAAAACGCCCGGGAGAUGAUCUGGCGCCUCAUCGUGGAGCAGACCAGCUCGAUGCUCGUGCUCUGGCUGGCCUGC-------------------------- ...((((((((((((((((.........)))).........(((((.....(((.((((......)))).)))...))))).))))))))))))-------------------------- ( -43.20, z-score = -2.15, R) >dp4.chrXL_group1e 8925113 94 - 12523060 GCAGUAGGCCAGCCAGCCGUAGAAGGCGCGCGAGAUGAUCUGCCGGCGCAUGGUGGAGCACACGAGCUCAAUGCUGGCCGUCUGACUGGCCUGG-------------------------- ....((((((((.(((..(((((...(((....).)).)))))((((((((....((((......)))).))))..)))).))).)))))))).-------------------------- ( -39.50, z-score = -0.24, R) >droPer1.super_22 403039 94 + 1688296 GCAAUAGGCCAGCCAGCCGUAGAAGGCGCGCGAGAUGAUCUGCCGGCGCAUGGUGGAGCACACGAGCUCAAUGCUGGCCGUCUGACUGGCCUGG-------------------------- ....((((((((.(((..(((((...(((....).)).)))))((((((((....((((......)))).))))..)))).))).)))))))).-------------------------- ( -39.60, z-score = -0.61, R) >droWil1.scaffold_180702 1537867 94 + 4511350 GCAAUAGGCCAACCAGCCAUAGAAAGCUCUCGAGAUUAUUUGACGCCGCAUGGUGGAGCAGACAAGCUCAAUGCUGGCCGUUUGACUGGCCUGU-------------------------- ...((((((((..(((((.......(((((((((....)))))((((....)))))))).....(((.....))))))....))..))))))))-------------------------- ( -29.80, z-score = -0.72, R) >droVir3.scaffold_12970 3147992 94 - 11907090 ACAGUAGGCCAGCCAGCCGUAGAAGGCGCGCGAGAUUAUCUGGCGACGCAUUGUGGAGCAGACCAGCUCAAUGCUGGCCGUUUGGCUGGCCUGU-------------------------- ....(((((((((((((..((.((.((((((.((.....)).))).))).)).))..)).(.(((((.....))))).)...))))))))))).-------------------------- ( -47.20, z-score = -3.58, R) >droMoj3.scaffold_6473 4001648 94 - 16943266 ACAGUAGGCCAGCCAGCCGUAGAAGGCGCGCGAGAUGAUCUGGCGACGCAUCGUGGAGCAGACCAGCUCAAUGCUGGCCGUUUGGCUGGCCUGU-------------------------- ....(((((((((((.(((..(...((((((.((.....)).))).))).)..)))(((.(.(((((.....)))))).)))))))))))))).-------------------------- ( -46.50, z-score = -2.99, R) >droGri2.scaffold_15081 3565247 94 + 4274704 ACAGUAGGCCAGCCAGCCGUAGAAGGCGCGCGAGAUGAUCUGGCGACGCAUCGUGGAGCAGACGAGCUCGAUGCUGGCCGUUUGGCUGGCCUGA-------------------------- ....((((((((((((((.......((((((.((.....)).))).)))..(((.((((......)))).)))..)))....))))))))))).-------------------------- ( -47.10, z-score = -3.08, R) >anoGam1.chr2R 28959350 120 + 62725911 ACAGUACGCCAGCCAGCCGUAGAAGGCGCGCGAAAUGAUCUGCCGCUUCAUCGUGCUGCAGACGAGCGCAAUGCUGGUGGAUUGGGUUGUCUGGGGCUGUUCGAGAACGAAACGAAAUAG ......(((((((..(((......)))((((....((.(((((.((........)).))))))).))))...)))))))..(((..((((((.((.....)).)).))))..)))..... ( -42.60, z-score = -0.47, R) >triCas2.ChLG6 8072506 79 - 13544221 GCAGUAAGCCAGCCAGCCGUAGAAAGCCCUAGAGAUGAUUUGGCGUUUCAUGGAAUCACAGACUGCUUGAAUACAAAUA----------------------------------------- (((((...........((((.(((((((..(((.....)))))).))))))))........))))).............----------------------------------------- ( -15.51, z-score = 0.01, R) >consensus ACAGUAGGCCAGCCAGCCGUAGAAGGCCCGCGAGAUGAUCUGCCGGCGCAUGGUGGAGCAGACCAGCUCAAUGCUGGUGGUCUGGCUGGCCUGG__________________________ ....(((((((((((((((((((.....(....)....)))).)).......((.((((......)))).))))))))........)))))))........................... (-20.46 = -21.28 + 0.82)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:44 2011