| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,298,943 – 20,299,054 |
| Length | 111 |
| Max. P | 0.848861 |

| Location | 20,298,943 – 20,299,054 |
|---|---|
| Length | 111 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 63.60 |
| Shannon entropy | 0.81921 |
| G+C content | 0.46495 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -9.63 |
| Energy contribution | -9.40 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
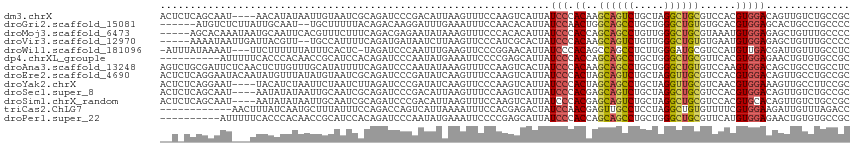
>dm3.chrX 20298943 111 - 22422827 ACUCUCAGCAAU----AACAUAUAAUUGUAAUCGCAGAUCCCGACAUUAAGUUUCCAAGUCAUUAUCCCACAAGCAGUCUGCUAGGCUGCGUCCACGUGGACAGUUGUCUGCCGC .......(((((----........)))))....((((((..((((.............)))............((((((.....))))))((((....)))).)..))))))... ( -27.82, z-score = -2.21, R) >droGri2.scaffold_15081 723449 107 - 4274704 ------AUGUCUCUUAUUGCAAU--UGCUUUUUACAGACAAGGAUUUGAAAUUUCCAACACAUUAUCCAACUGGCAGCCUGCUGGGCUGUGUGCACGUGGAGCACUGCCUGCCCC ------.(((((......((...--.)).......)))))(((...((....(((((.((((....((....))(((((.....)))))))))....)))))))...)))..... ( -23.52, z-score = 1.19, R) >droMoj3.scaffold_6473 12957810 110 + 16943266 -----AGCACAAAUAAUGCAAUUCACGUUUCUUUCAGACGAGAAUAUAAAGUUUCCCACACAUUAUCCCACCAGCAGCCUGUUGGGCUGCGUAAAUGUGGAGAGCUGUUUGCCCC -----............(((((((.(((((.....))))).))))....((((..(((((..((((.......((((((.....)))))))))).)))))..))))...)))... ( -29.51, z-score = -2.10, R) >droVir3.scaffold_12970 8773545 108 + 11907090 -----AAAAUAAUUGAUUACGUU--UGCCAUUUUCAGAUGAUAAUCUUAAGUUCCCAUCGCACUAUCCCACAAGCAGUCUGUUGGGCUGUGUGAAUGUGGAGAGCUGUUUGCCCC -----.........(((((((((--((.......)))))).)))))...((((((((((((((...((((((.......)).))))..))))))...))).)))))......... ( -28.30, z-score = -1.88, R) >droWil1.scaffold_181096 10812303 110 + 12416693 -AUUUAUAAAAU---UUCUUUUUUAUUUCACUC-UAGAUCCCAAUUUGAAGUUCCCGGAACAUUAUCCCACAGCCAGCCUCUUGGGAUGCGUCCAUGUUGACGAUUGUUUGCCUC -........(((---(((.........((....-..)).........))))))....(((((.(((((((.((.......)))))))))((((......))))..)))))..... ( -15.87, z-score = 0.68, R) >dp4.chrXL_group1e 3614080 105 + 12523060 ----------AUUUUUCACCCACAACCGCAUCCACAGAUCCCAAUAUGAAAUUCCCCGAGCAUUAUCCCACCAGCAGCCUGCUGGGCUGCGUUCACGUGGAGAACUGUGUGCCGC ----------.................(((..(((((.(((((...((((.......((......))......((((((.....)))))).))))..))).)).))))))))... ( -28.30, z-score = -1.77, R) >droAna3.scaffold_13248 3500574 115 - 4840945 AGUCUGCGAUUCUCAACUCUUGUUUGCAUAUUUUCAGAUCCCAAUAUAAAGUUUCCAAGUCACUAUCCCACAAGCAGCCUGCUGGGCUGUGUCCAAGUGGACAGCUGCCUGCCUC .(((..(((((...((((.(((((((...(((....)))..))).))))))))....))))............((((((.....))))))......)..))).((.....))... ( -21.50, z-score = 1.27, R) >droEre2.scaffold_4690 16906040 115 - 18748788 ACUCUCAGGAAUACAAUAUGUUUAUAUGUAAUCGCAGAUCCCGAUAUCAAGUUUCCAAGUCAUUAUCCCACUAGCAGUCUGCUAGGUUGCGUCCACGUGGACAGUUGCCUGCCGC .....((((....((((.((((((..((....(((((.....((((.................))))...(((((.....))))).)))))..))..)))))))))))))).... ( -23.63, z-score = 0.06, R) >droYak2.chrX 19524867 111 - 21770863 ACUCUCAGGAAU----UACAUCUAAUUCUAAUCUUAGAUCCCGAUAUCAAGUUCCCAAGUCAUUAUCCCACUAGCAGCCUGCUAGGUUGCGUCAACGUGGAAAGUUGCCUUCCGC ...((..(((((----(..(((....((((....))))....)))....))))))..))..............((((((.....))))))......((((((.......)))))) ( -22.50, z-score = -1.13, R) >droSec1.super_8 2492543 111 - 3762037 ACUCUCAGCAAU----AAUAUAUAAUUGCAAUCGCAGAUCCCGACAUUAAGUUUCCAAGUCAUUAUCCCACGAGCAGUCUGCUAGGCUGCGUCCACGUGGACAGUUGUCUGCCGC .......(((((----........)))))....((((((..((((.............)))......(((((.((((((.....)))))).....)))))...)..))))))... ( -31.02, z-score = -2.66, R) >droSim1.chrX_random 5279435 111 - 5698898 ACUCUCAGCAAU----AAUAUAUAAUUGCAAUCGCAGAUCCCGACAUUAAGUUUCCAAGUCAUUAUCCCACGAGCAGUCUGCUAGGCUGCGUCCACGUGCACAGUUGUCUGCCGC .......(((((----........)))))....((((((..((((.............))).......((((.((((((.....)))))).....))))....)..))))))... ( -27.02, z-score = -1.78, R) >triCas2.ChLG7 9226025 103 + 17478683 ------------AACUUUAUCAAUGCUUUAUUUCCAGACCAGUCAUUAAAAUUUCCACGAGACUAUCCAACGAGUUGCCUCCUAGGCUGUGUUUUCGUGGAAGAUUGUUUAGACC ------------..((....((((............((....)).......(((((((((((((........))))(((.....))).......)))))))))))))...))... ( -19.90, z-score = -0.78, R) >droPer1.super_22 1301773 105 - 1688296 ----------AUUUUUCACCCACAACCGCAUCCACAGAUCCCAAUAUGAAAUUCCCCGAGCAUUAUCCCACCAGCAGCCUGCUGGGCUGCGUUCAUGUGGAGAACUGUGUGCCGC ----------.................(((..(((((.(((((.((((((.......((......))......((((((.....)))))).))))))))).)).))))))))... ( -30.60, z-score = -2.41, R) >consensus __U_UCAGAAAU___AUACAUAUAAUUGUAUUCGCAGAUCCCAAUAUGAAGUUUCCAAGUCAUUAUCCCACAAGCAGCCUGCUGGGCUGCGUCCACGUGGACAGUUGUCUGCCGC .................................................................(((.((..((((((.....))))))......))))).............. ( -9.63 = -9.40 + -0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:39 2011