
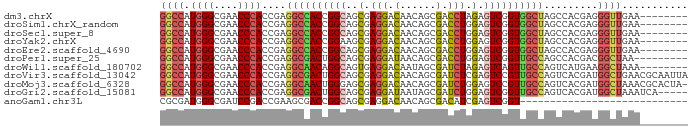
| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,282,502 – 20,282,582 |
| Length | 80 |
| Max. P | 0.795866 |

| Location | 20,282,502 – 20,282,582 |
|---|---|
| Length | 80 |
| Sequences | 11 |
| Columns | 88 |
| Reading direction | forward |
| Mean pairwise identity | 82.11 |
| Shannon entropy | 0.38398 |
| G+C content | 0.63983 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -24.37 |
| Energy contribution | -24.30 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.795866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
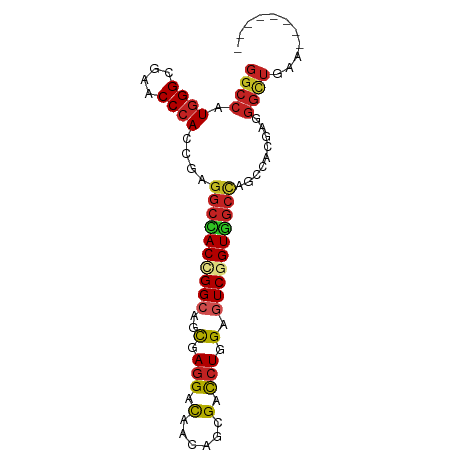
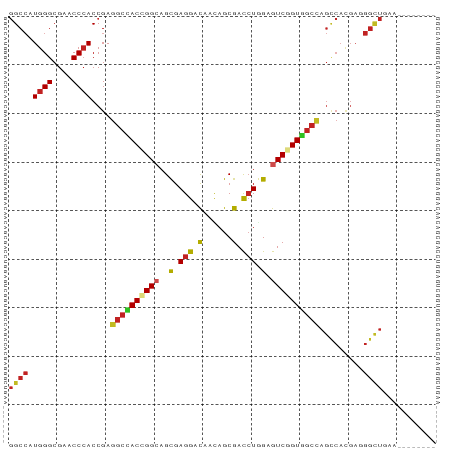
>dm3.chrX 20282502 80 + 22422827 GGCCAUGGGCGAACCCACCGAGGCCACCGGCAGCGAGGACAACAGCGACCUAGAGUCGGUGGCUAGCCACGAGGGUUGAA-------- (((..((((....))))....((((((((((....(((.(......).)))...)))))))))).)))............-------- ( -31.00, z-score = -1.14, R) >droSim1.chrX_random 5272287 80 + 5698898 GGCCAUGGGCGAACCCACCGAGGCCACCGGCAGCGAGGACAACAGCGACCUGGAGUCGGUGGCUAGCCACGAGGGUUGAA-------- (((..((((....))))....((((((((((..(.(((.(......).))).).)))))))))).)))............-------- ( -32.20, z-score = -0.98, R) >droSec1.super_8 2475902 80 + 3762037 GGCCAUGGGCGAACCCACCGAGGCCACCGGCAGCGAGGACAACAGCGACCUGGAGUCGGUGGCUAGCCACGAGGGUUGAA-------- (((..((((....))))....((((((((((..(.(((.(......).))).).)))))))))).)))............-------- ( -32.20, z-score = -0.98, R) >droYak2.chrX 19508341 80 + 21770863 GGCCAUGGGCGAACCCACCGAGGCCACCGGAAGCGAGGACAACAGCGACCUGGAGUCGGUGGCUAGCCACGAGGGUUGAA-------- (((..((((....))))....(((((((((...(.(((.(......).))).)..))))))))).)))............-------- ( -31.00, z-score = -0.97, R) >droEre2.scaffold_4690 10350303 80 + 18748788 GGCCAUGGGCGAACCCACCGAGGCCACCGGCAGCGAGGACAACAGCGACCUGGAGUCGGUGGCUAGCCACGAGGGUUGAA-------- (((..((((....))))....((((((((((..(.(((.(......).))).).)))))))))).)))............-------- ( -32.20, z-score = -0.98, R) >droPer1.super_25 335619 79 + 1448063 GGCCAUGGGCGAACCCACCGAGGCGACUGGCAGCGAGGAUAACAGCGACCUGGAGUCGGUUGCCAGCCACGACGGCUAA--------- ((((.((((....)))).((.(((...(((((((...(((..(((....)))..))).)))))))))).))..))))..--------- ( -31.60, z-score = -1.51, R) >droWil1.scaffold_180702 4424473 80 + 4511350 GGCCAUGGGCGAACCCACCGAGGCAACAGGCAGUGAGGACAAUAGCGAUCUAGAGUCAGUUGCCAGUCAUGAAGGCUAAA-------- ((((.((((....))))....((((((.(((....(((.(......).)))...))).)))))).........))))...-------- ( -23.80, z-score = -0.99, R) >droVir3.scaffold_13042 2136221 88 - 5191987 GGCCAUGGGCGAACCCACCGAGGCGACUGGCAGCGAGGACAACAGCGAUCUCGAGUCCGUUGCCAGUCACGAUGGCUGAACGCAAUUA (((((((((....)))......(.((((((((((..((((.....((....)).)))))))))))))).).))))))........... ( -35.90, z-score = -2.42, R) >droMoj3.scaffold_6328 609415 87 + 4453435 GGCCAUGGGCGAACCCACCGAGGCAACUGGGAGCGAGGACAACAGCGAUCUGGAGUCCGUUGCCAGUCACGAUGGCUAAACGCACUA- (((((((((....))).((.((....)).)).((((((((..(((....)))..)))).))))........))))))..........- ( -31.60, z-score = -1.32, R) >droGri2.scaffold_15081 3185384 83 + 4274704 GGCCAUGGGCGAACCCACCGAGGCGACUGGCAGCGAGGAUAAUAGCGAUCUGGAGUCGGUUGCCAGUCACGAUGGCUAAAUCA----- (((((((((....)))......(.((((((((((...(((..(((....)))..))).)))))))))).).))))))......----- ( -32.40, z-score = -1.94, R) >anoGam1.chr3L 33174858 60 + 41284009 CGCGAUGGGCGAUCCGACCGAAGCGACCGGCAGCGAGGACAACAGCGACAUCGAGUCGGU---------------------------- (((.....)))..((((((((.((.....)).((..(.....).))....))).))))).---------------------------- ( -16.60, z-score = 0.79, R) >consensus GGCCAUGGGCGAACCCACCGAGGCCACCGGCAGCGAGGACAACAGCGACCUGGAGUCGGUGGCCAGCCACGAGGGCUGAA________ ((((.((((....))))....((((((((((..(.(((.(......).))).).)))))))))).........))))........... (-24.37 = -24.30 + -0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:35 2011