| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,243,042 – 20,243,125 |
| Length | 83 |
| Max. P | 0.782941 |

| Location | 20,243,042 – 20,243,125 |
|---|---|
| Length | 83 |
| Sequences | 10 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 94.51 |
| Shannon entropy | 0.11827 |
| G+C content | 0.35423 |
| Mean single sequence MFE | -18.40 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
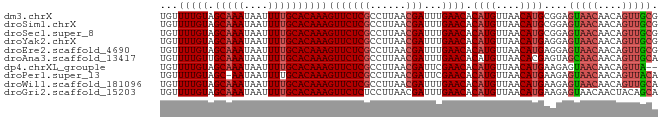
>dm3.chrX 20243042 83 - 22422827 UGUUUUGUAGCAAAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUUGAACACAUGUUAACAUGCGGAGUAACAACAGUUGCG ...(((((.(((((....))))))))))(((((((......)))...)))).((((....))))....(((((....))))). ( -18.80, z-score = -1.08, R) >droSim1.chrX 15518166 83 - 17042790 UGUUUUGUAGCAAAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUUGAACACAUGUUAACAUGCGGAGUAACAACAGUUGCG ...(((((.(((((....))))))))))(((((((......)))...)))).((((....))))....(((((....))))). ( -18.80, z-score = -1.08, R) >droSec1.super_8 2430019 83 - 3762037 UGUUUUGUAGCAAAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUUGAACACAUGUUAACAUGCGGAGUAACAACAGUUGCG ...(((((.(((((....))))))))))(((((((......)))...)))).((((....))))....(((((....))))). ( -18.80, z-score = -1.08, R) >droYak2.chrX 19474506 83 - 21770863 UGUUUUGUAGCAAAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUUGAACACAUGUUAACAUGAGGAGUAACAACAGUUGCG ..((((((.(((((....)))))))))))((((((..((((((...((....))))))))..))))))(((((....))))). ( -19.60, z-score = -1.70, R) >droEre2.scaffold_4690 10315559 83 - 18748788 UGUUUUGUAGCAAAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUUGAACACAUGUUAACAUGAGGAGUAACAACAGUUGCG ..((((((.(((((....)))))))))))((((((..((((((...((....))))))))..))))))(((((....))))). ( -19.60, z-score = -1.70, R) >droAna3.scaffold_13417 1518545 83 - 6960332 UGUUUUGUUGCAAAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUUGAACACAUGUUAACACGAGUAGCAACAACAGUUGCA ..((((((.(((((....)))))))))))..((((..((((((...((....))))))))..))))..(((((....))))). ( -20.60, z-score = -1.73, R) >dp4.chrXL_group1e 7035093 81 - 12523060 UGUUUUGUAGCAAAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUCGAACACAUGUUAACAUGAAGAGUAACAACAGUUA-- ...(((((.(((((....))))))))))(((((((......)))...)))).((((....)))).................-- ( -15.10, z-score = -1.08, R) >droPer1.super_13 774434 82 + 2293547 UGUUUUGUAGC-AAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUCGAACACAUGUUAACAUGAAGAGUAACAACAGUUACA ...(((((.((-((.....)))))))))(((((((......)))...)))).((((....))))....(((((....))))). ( -18.60, z-score = -2.28, R) >droWil1.scaffold_181096 9284113 83 - 12416693 UGUUUUGUAGCAAAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUUGAACACAUGUUAACAUGAAGAGUAACAACAGUUGCA ...(((((.(((((....))))))))))(((((((......)))...)))).((((....))))....(((((....))))). ( -19.50, z-score = -1.87, R) >droGri2.scaffold_15203 5907247 83 - 11997470 UGUUUUGUAGCAAAUAAUUUUGCACAAAGUUCUCUCCUUAACGAUUUGAACACAUGUUAACAUGAAGAGUAACAACUACAGCA ...(((((.(((((....))))))))))(((((((..((((....))))...((((....)))).)))).))).......... ( -14.60, z-score = -0.80, R) >consensus UGUUUUGUAGCAAAUAAUUUUGCACAAAGUUCUCGCCUUAACGAUUUGAACACAUGUUAACAUGAGGAGUAACAACAGUUGCG ...(((((.(((((....))))))))))(((((((......)))...)))).((((....))))....(((((....))))). (-17.05 = -17.50 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:30 2011