| Sequence ID | dm3.chrX |
|---|---|
| Location | 20,238,022 – 20,238,139 |
| Length | 117 |
| Max. P | 0.783377 |

| Location | 20,238,022 – 20,238,139 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.36 |
| Shannon entropy | 0.36738 |
| G+C content | 0.58856 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -27.38 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 20238022 117 + 22422827 CGGCUGUCAGGUGUCGCCGCACUCCGACUCAAAGUUAUGUGUUAUCAUUCCGCCGAGGUCGGCGUUGACUGGCUCCACGUGGGCCGCCAGUACCUUCCAAAGGCAUAACACUUCAAA .....(((.(((((....)))))..))).....(((((((.((.......(((((....)))))...(((((((((....)))..))))))........)).)))))))........ ( -36.20, z-score = -0.32, R) >droSec1.super_8 2425137 116 + 3762037 CGGCUGUCAGGUGUCGCCGCACUCCGACUCAAAGUUAUGUGUUAUCAUUCCGCCG-GGUCGGCGUUGACUGGCUCCACGUGAGCCGCCAGUACCCCCCAAAGGUACUACACUUCAUC .(((.((((.(((..((((.((.(((((((...(..(((......)))..)...)-)))))).))....))))..))).))).).)))((((((.......)))))).......... ( -40.90, z-score = -1.99, R) >droEre2.scaffold_4690 10310687 91 + 18748788 CGGCUGUCAGGUGUCGCCGCGCUCCGGCUCAAAGUUAUGUGUUAUCAUUCCGCCGGGCCCGGCAUGGAGUAGCUCCAUGUGAGCCCUCAUC-------------------------- .((((..........((((.((.(((((.....(..(((......)))..)))))))).))))((((((...))))))...))))......-------------------------- ( -32.90, z-score = -0.91, R) >consensus CGGCUGUCAGGUGUCGCCGCACUCCGACUCAAAGUUAUGUGUUAUCAUUCCGCCG_GGUCGGCGUUGACUGGCUCCACGUGAGCCGCCAGUACC__CCAAAGG_A__ACACUUCA__ .((((..((.(((..(((((((...(((.....)))..))))..(((...(((((....))))).)))..)))..))).))))))................................ (-27.38 = -27.50 + 0.12)


| Location | 20,238,022 – 20,238,139 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.36 |
| Shannon entropy | 0.36738 |
| G+C content | 0.58856 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -26.13 |
| Energy contribution | -27.80 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
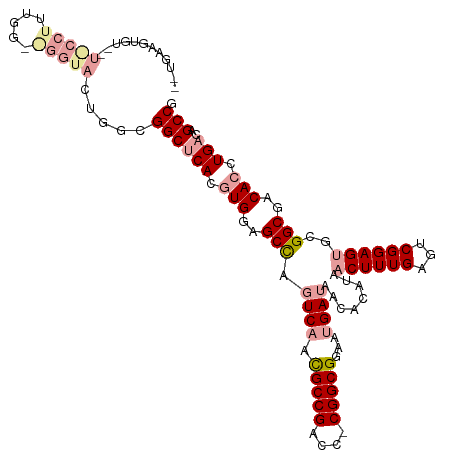
>dm3.chrX 20238022 117 - 22422827 UUUGAAGUGUUAUGCCUUUGGAAGGUACUGGCGGCCCACGUGGAGCCAGUCAACGCCGACCUCGGCGGAAUGAUAACACAUAACUUUGAGUCGGAGUGCGGCGACACCUGACAGCCG ......(((((((((((.....))))((((((..((.....)).))))))...(((((....))))).....)))))))..........(((((.(((......))))))))..... ( -40.60, z-score = -1.15, R) >droSec1.super_8 2425137 116 - 3762037 GAUGAAGUGUAGUACCUUUGGGGGGUACUGGCGGCUCACGUGGAGCCAGUCAACGCCGACC-CGGCGGAAUGAUAACACAUAACUUUGAGUCGGAGUGCGGCGACACCUGACAGCCG ..........(((((((.....)))))))((((((((.....))))).((((.((((((((-((((.......................))))).)).))))).....)))).))). ( -43.50, z-score = -1.43, R) >droEre2.scaffold_4690 10310687 91 - 18748788 --------------------------GAUGAGGGCUCACAUGGAGCUACUCCAUGCCGGGCCCGGCGGAAUGAUAACACAUAACUUUGAGCCGGAGCGCGGCGACACCUGACAGCCG --------------------------.....((((((.(((((((...)))))))..))))))((((..(((......)))..).....((((.....))))...........))). ( -34.30, z-score = -2.41, R) >consensus __UGAAGUGU__U_CCUUUGG__GGUACUGGCGGCUCACGUGGAGCCAGUCAACGCCGACC_CGGCGGAAUGAUAACACAUAACUUUGAGUCGGAGUGCGGCGACACCUGACAGCCG ............(((((.....))))).....((((((.(((..(((.((((.(((((....)))))...))))........((((((...))))))..)))..))).)))..))). (-26.13 = -27.80 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:03:28 2011